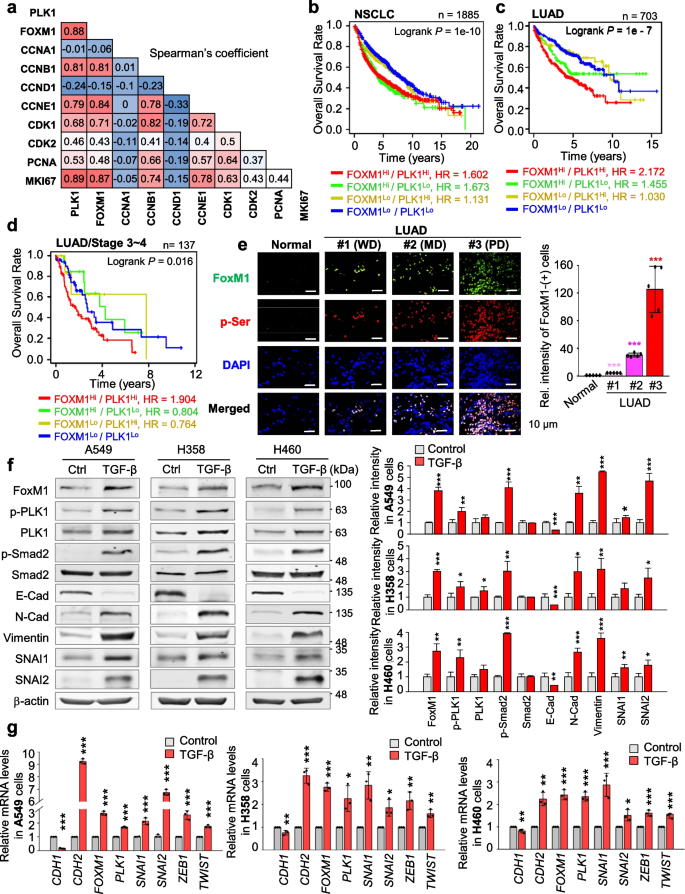
Concurrent upregulation of FoxM1 and PLK1 correlated with poor survival of lung adenocarcinoma (LUAD) patients
The majority of KEGG pathways in active PLK1-induced invasive NSCLC is related to ECM-adhesion and cell cycle-related factors [29] (Fig. S2a). Because proliferation is closely connected with metastasis [31], we investigated proliferating factors related to major metastatic drivers in PLK1-driven invasiveness [29]. Under conditions of low E-cadherin (CDH1) and high N-cadherin (CDH2), TGFB1, and TGFB2 in active PLK1-expressed NSCLC cells, the expression of cell cycle factors including FOXM1, CCNB1, CCNE1, and MKI67 was highly correlated with PLK1 expression (Fig. S2b). In mRNA expression, the correlation between PLK1 and FOXM1 (Spearman: 0.88, p = 1.68e-55; Pearson: 0.86, p = 3.22e-51) or MKI67 (Spearman: 0.89, p = 3.67e-59; Pearson: 0.89, p = 1.13e-59) was positive compared with those of others, including CCNA1 and CCND1, extracted from the cBio-Portal analysis in NSCLC patients (Fig. 1a, Fig. S2c-d, Table S5). Notably, both PLK1 and FoxM1 are regulators for EMT as well as mitosis [8,9,10]. For their clinical relationship, the overall survival (OS) corresponding to the expression level of each factor in NSCLC or LUAD was observed using a Kaplan–Meier (KM) plot database [41]. The clinical features of lung cancer patients were presented (Table S6). The OS with high FOXM1/PLK1 expression was significantly shorter than that with low FOXM1/PLK1 expression (NSCLC, n = 1885, HR = 1.602, log-rank P = 1e-10; LUAD, n = 703, HR = 2.172, log-rank P = 1e-7) (Fig. 1b-c, Table S6-S7). Survival until first progression (FPS) rates of patients with NSCLC or LUAD were correlated with FOXM1/PLK1 expression, similar to the correlation with OS (NSCLC, n = 982, HR = 1.75, log-rank P = 1.2e-08; LUAD, n = 461, HR = 2.58, log-rank P = 5.3e-09; Fig. S2e-f). However, the OS or FPS rates of patients with lung squamous cell cancer (LUSQ) were not significant (Fig. S2g). In a clinical analysis of metastatic NSCLC or LUAD patients at TNM stage N1, the OS of patients with high FOXM1/PLK1 expression was shorter than for those with low FOXM1/PLK1 (Fig. S2h). In addition, clinical analysis of 137 advanced LUAD patients at stages 3–4 showed shorter OS rates of advanced patients with high FOXM1/PLK1 expression than of those with low expression (Fig. 1d, n = 137, HR = 1.904, log-rank P = 0.016; Table S7). These data indicate that high expression of FoxM1 and PLK1 occurs concurrently with a poor prognosis for patients with primary and advanced NSCLC, especially LUAD.
Concurrent upregulation of FoxM1 and PLK1 is correlated with poor survival of LUAD patients. a Analysis of Spearman’s coefficient for the correlations between cell cycle-regulatory factors including PLK1, FOXM1, CCNA1, CCNB1, CCND1, CCNE1, CDK1, CDK2, PCNA, and MKI67. b-d The overall survival (OS) of patients with non-small lung cancer (n = 1885) (b), lung adenocarcinoma (LUAD) (n = 703) (c), and stages 3–4 of LUAD (n = 137) (d) were analyzed according to PLK1 and FOXM1 expression levels using KM PLOTTER. High (Hi) and low (Lo) were generated by separating patients according to expression at the median cut-off. e Using human lung tissues from LUAD patients and normal individual, immunohistochemistry analysis was displayed with anti-FoxM1 (Green) and anti-p-Serinie (Red) antibodies. The relative intensity of cells that exhibited positive FoxM1 was analyzed and plotted. WD, well differentiated (grade 1); MD, moderately differentiated (grade 2); PD, poorly differentiated (Grade 3), n > 5000. *p < 0.05; **p < 0.01; ***p < 0.001. Data are presented as mean ± SD. f and g A549, NCI-H358 (H358), and NCI-H460 (H460) NSCLC cells treated with 5 ng/mL of TGF-β for 48 h. f Immunoblotting was performed to measure the expression and phosphorylation of PLK1 using specific antibodies for FoxM1, PLK1, p-PLK1T210, p-Smad2S465/S467, Smad2, E-cadherin, N-cadherin, vimentin, SNAI1, SNAI2, and β-actin in A549, NCI-H358 (H358), and NCI-H460 (H460) cells (left panel). The relative band intensities for FoxM1, p-PLK1T210, PLK1, p-Smad2.S465/S467, Smad2, E-cadherin, N-cadherin, vimentin, SNAI1, and SNAI2 were quantified using LI-COR Odyssey software (right panel). g qRT-PCR was performed for CDH1, CDH2, FOXM1, PLK1, SNAI1, SNAI2, ZEB1, and TWIST expression in A549 (left panel), NCI-H358 (middle panel), and NCI-H460 (right panel) cells. Data are presented as mean ± SD of at least three independent experiments (significantly different from the experimental control). *p < 0.05; **p < 0.01; ***p < 0.001
To understand FOXM1 expression in metastatic LUAD, data from TCGA were analyzed by stage in normal and tumor tissue, constructing a heatmap based on the degree of expression of FOXM1 (Fig. S2i). In genomic analysis, FOXM1 expression was much higher in the advanced stages 2–4 (81%; 22 tumors/27 total) than those of primary tumor stage 1 (70%; 17 tumors/24 total). The expression pattern of FOXM1 was like that of other proliferation factors including PLK1, CCNB1, and MKI67. SNAI1, a mesenchymal marker, was upregulated compared with normal tissues in stage 2–4 LUAD patients having high FOXM1 (Fig. S2i). Accordingly, the concurrent high expression of FoxM1 and PLK1 showed a high correlation with survival of primary and advanced NSCLC patients, particularly in LUAD but not in LUSQ.
To explore the phosphorylation and expression of FoxM1 in LUAD patients, we carried out immunohistochemistry on tissues from LUAD patients (Super Bio Chips, Korea), categorized by their tumor grade (Table S8). These tissues were subjected to staining with anti-FoxM1 and anti-p-Serine antibodies. FoxM1 and p-Serine were co-stained at the same location within LUAD tissues. Importantly, we observed a notable increase in the levels of both FoxM1 and p-Serine in LUAD patients compared to those from a healthy individual with normal lung tissues (Fig. 1e, Table S8). Furthermore, these elevated levels were even more pronounced in advanced LUAD tissues when compared to low-grade LUAD tissues.
FoxM1 is phosphorylated by PLK1 through direct interaction in TGF-β -induced EMT
To investigate the functions of FoxM1 and PLK1 in the EMT, we observed the expression of FoxM1 and PLK1 in primary A549 and metastatic NCI-H358, NCI-H1299, and NCI-H460 NSCLC cells. Compared with normal lung epithelial BEAS-2B cells, the expression of FoxM1 and PLK1 was higher in NSCLC cells (Fig. S3a). The level of active PLK1 phosphorylated at Thr210 was higher in NSCLC cells than in normal BEAS-2B cells. Using previously published microarray data of lung cancer (GSE 114761), FoxM1 and PLK1 were upregulated in TGF-β-induced EMT in A549, NCI-H522, NCI-H1944, and NCI-H2122 cells (Fig. S3b). Additionally, changes of FoxM1 and PLK1 were observed in TGF-β-treated A549, NCI-H358, and NCI-H460 cells. Treatment with TGF-β, an inducer of the EMT, increased the expression of CDH2, a mesenchymal marker, but decreased the level of CDH1, an epithelial index, relative to those of control cells (Fig. 1f-g). Under these conditions, immunoblot analysis revealed that FoxM1 proteins increased approximately 3.8, 3.1, and 2.8 times in A549, NCI-H358, and NCI-H460 cells, respectively. This increase in FoxM1 levels coincided with the upregulation of p-Smad2, a downstream factor of TGF-β signaling pathway, induced by TGF-β treatment (Fig. 1f, Fig. S3c). In response to TGF-β treatment, the levels of total and the active form of PLK1 phosphorylated at Thr210 increased in TGF-β-treated NSCLC cells (Fig. 1f, Fig. S3c). qRT-PCR showed that PLK1 and FOXM1 levels increased by a factor of 2–3 compared with a control in TGF-β-treated NSCLC cells when the transcriptional factors for EMT including SNAI1, SNAI2, ZEB1, and TWIST increased (Fig. 1g). Therefore, active PLK1 and FoxM1 are concurrently upregulated in TGF-β-induced EMT of NSCLC.
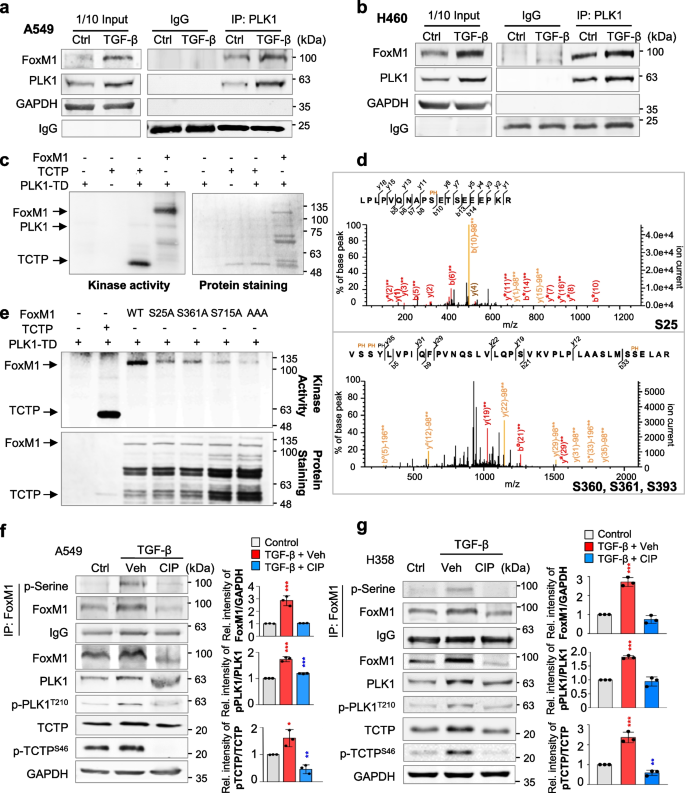
Because of upregulation of active PLK1 and FoxM1 in TGF-β-induced EMT (Fig. 1) and their functions as positive EMT regulators [9, 10, 36, 37], we hypothesized that FoxM1 and PLK1 work cooperatively in TGF-β-induced EMT. For this, immunoprecipitation assays were performed to observe their interaction in TGF-β-induced EMT. In TGF-β-treated A549 cells and NCI-H460 cells, endogenous PLK1 interacted with endogenous FoxM1 (Fig. 2a-b). Exogenously expressed FoxM1 also interacted with PLK1 in TGF-β-treated A549 cells (Fig. S4a). Thus, PLK1 and FoxM1 interact directly during TGF-β-induced EMT.
TGF-β-treated EMT results in phosphorylation of FoxM1 by PLK1 by direct interaction. a, b A549 (a) and NCI-H460 (b) were treated with TGF-β (5 ng/mL) for 48 h. Immunoprecipitation of cell lysates was performed with normal IgG or anti-PLK1 antibody and then immunoblotting was performed with anti-FoxM1 antibody. c An in vitro kinase assay was performed with an active version of PLK1 with T210D (PLK1-TD), radioactive ATP, and purified GST-FoxM1. GST-tagged TCTP was used as the positive control. d In LC–MS/MS analysis, possible phosphorylation residues of FoxM1 by PLK1 were newly detected at the S25, S360, S361, and S393 sites. e, Purified GST-tagged wild-type, S25A, S361A, S715A, and S25/S361/S715A (AAA) FoxM1 mutants were used for a PLK1 kinase assay with radioactive ATP. f, g Phosphorylation of FoxM1 in A549 (f) and NCI-H358 (g) cells treated with TGF-β for 48 h. Treatment with calf intestinal alkaline phosphatase (CIP) reduced the phosphorylation of FoxM1 and PLK1 in TGF-β-induced EMT. Immunoprecipitation was performed with anti-normal IgG (Fig. S4d-e) or anti-FoxM1 antibody, and then immunoblotting was conducted with anti-p-Serine antibodies. Immunoblotting was performed for FoxM1, PLK1, p-PLK1T210, TCTP, and p-TCTPS46 using specific antibodies. TCTP was used as a positive control of the PLK1 substrate. Data are presented as mean ± SD of at least three independent experiments (significantly different from the experimental control). *p < 0.05; **p < 0.01; ***p < 0.001
Since active PLK1 drives the EMT, to explore phosphorylation of sites of FoxM1 by PLK1 for EMT, in vitro PLK1 kinase assays and LC–MS/MS analyses were performed. In the tests, PLK1 strongly phosphorylated FoxM1 as much as did TCTP, a positive control (Fig. 2c). LC–MS/MS analysis illustrated newly predicted phosphorylation sites at the Ser25, Ser360, Ser361, and Ser393 residues (Fig. 2d). Non-phosphomimetic alanine substitutes of FoxM1 at these newly predicted phosphorylation sites and at previously found phosphorylation residues (Ser715 and Ser724) [28] were generated using site-directed mutagenesis. The in vitro PLK1 kinase assay showed that the band intensity of non-phosphomimetic alanine-single mutants at Ser25, Ser361, and Ser715 residues was markedly reduced compared with that of wild-type FoxM1 (Fig. 2e, Fig. S4b). Triple mutant (S25A/S361A/S715A; AAA) was barely phosphorylated by PLK1 in vitro PLK1 kinase assay (Fig. 2e, Fig. S4c). In addition to the previously found Ser715 residue, Ser25 and Ser361 residues of FoxM1 were potent phosphorylation sites by PLK1. To examine whether FoxM1 phosphorylation depends on the EMT, A549 and NCI-H358 cells were treated with TGF-β (Fig. 2f-g). Phosphatase treatment reduced the levels of p-FoxM1Ser, p-PLK1T210, and p-TCTPS46 and retarded the shifted bands of PLK1, TCTP, and FoxM1, which were upregulated by TGF-β treatment (Fig. 2f-g, Fig. S4d-e), implying that phosphorylation of FoxM1 by PLK1 occurs during the EMT in both primary A549 and metastatic NCI-H460 LUAD cells. These phosphorylation residues are evolutionarily conserved in several species (Fig. S4f).
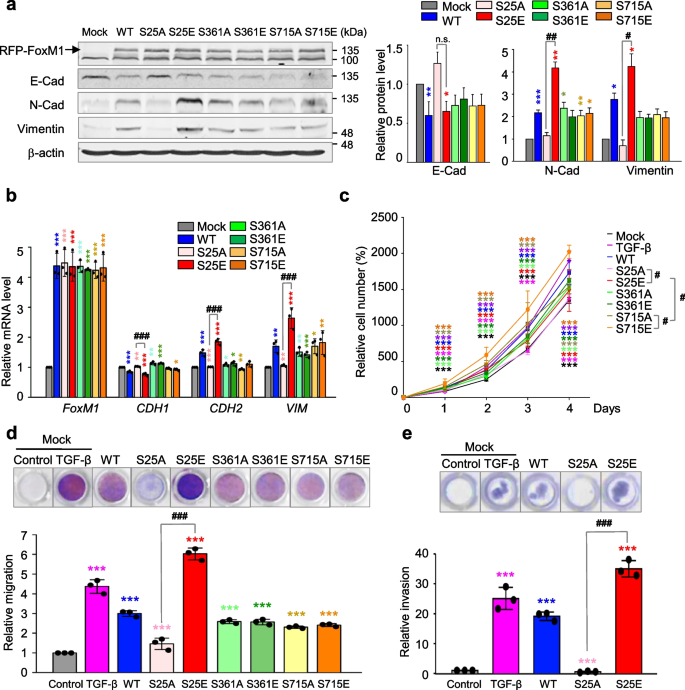
Phosphorylation of FoxM1 at Ser25 facilitates cell migration and invasiveness but not cell proliferation
We then studied functions of p-FoxM1 for the EMT. The phosphomimetics and non-phosphomimetics of FoxM1 were expressed in primary LUAD A549 cells, using a doxycycline-inducible expression system and site-specific mutagenesis to replace the serine residues with alanine for non-phosphomimetics and with glutamic acid for phosphomimetics. Under conditions with similar expression of different versions of FoxM1, the expression of S25E FoxM1 (FoxM1S25E) increased protein and mRNA levels of mesenchymal markers N-cadherin and vimentin, but the expression of expressing S25A FoxM1 (FoxM1S25A) did not (Fig. 3a-b, Fig. S1), indicating that expression of FoxM1S25E induces the mesenchymal transition from epithelial cells. However, cells expressing phosphomimetics and non-phosphomimetics at Ser361 and Ser715 did not show differences in the levels of N-cadherin or vimentin. To understand the proliferation effects of each version of FoxM1, a cell proliferation assay was performed (Fig. 3c). A549 cells expressing FoxM1S715E showed the highest proliferation effects, as reported previously for mitosis [44]. However, cells expressing phosphomimetics and non-phosphomimetics at the Ser25 and Ser361 residues were similar in terms of proliferation rate, indicating that phosphorylation at Ser25 and Ser361 of FoxM1 does not foster cell proliferation.
Phosphorylation of FoxM1 at Ser25 facilitates cancer cell migration and invasiveness but not cell proliferation. RFP-tagged wild-type (WT) FoxM1 and S25A, S25E, S361A, S361E, S715A, and S715E mutants were expressed in A549 cells. A549 cells were treated with doxycycline to express RFP-tagged FoxM1. a Immunoblotting was performed using specific antibodies for RFP, N-cadherin (N-Cad), E-cadherin (E-Cad), vimentin, and β-actin (left panel). The band intensity values were quantified using LI-COR Odyssey software, normalized, and plotted (right panel). b qRT-PCR was performed for FOXM1, CDH1, CDH2, and VIM in A549 cells expressing wild-type or mutants FoxM1. *p < 0.05; **p < 0.01; ***p < 0.001; (n = 3). Data are presented as mean ± SD. c Cell proliferation assay was performed (n = 3). Data are presented as mean ± SD of at least three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001 compared with experimental control; #p < 0.05 compared with indicated groups of cells. d Cells expressing wild-type or mutants of FoxM1 were subjected to a Transwell migration assay. As a positive control for migration, cells were treated with TGF-β. Three days after seeding, the cells on the bottom surface were stained with 0.05% crystal violet dye. Images of the Transwell cell migration assay were collected and analyzed with an Odyssey infrared imaging system and plotted. e An invasion assay was performed using A549 cells expressing wild-type or mutants of FoxM1. Seven days after seeding, the cells that invaded the bottom surface were stained with 0.05% crystal violet dye, and the relative absorbance was plotted. Data are presented as mean ± SD of at least three independent experiments (significantly different from the experimental control). *p < 0.05; **p < 0.01; ***p < 0.001 compared with experimental control. #p < 0.05; ##p < 0.01; ###p < 0.001 compared with S25A FoxM1
Next, we explored phosphorylation sites of FoxM1 related to the EMT. For this, a Transwell cell migration assay and an inverted invasion assay were performed in A549 cells expressing phosphomimetic and non-phosphomimetic FoxM1 (Fig. 3d-e, Fig. S1). TGF-β treatment increased cell migration and invasiveness approximately 5- and 25-fold compared with the control, respectively. The expression of FoxM1S25E upregulated cell migration and invasiveness approximately 6- and 35-fold compared with the mock system, respectively. Cell migration and invasiveness were lower in cells expressing FoxM1S25A than FoxM1WT. However, migration of cells expressing phosphomimetics and non-phosphomimetics at Ser361 and Ser715 residues was similar (Fig. 3d), indicating no relation of the function of phosphorylation at Ser361 and Ser715 of FoxM1 with the EMT. These patterns were consistent in LUAD HCC827 cells expressing FoxM1S25E (Fig. S5a) with higher CDH2 and lower CDH1 than those of mock or FoxM1WT and higher invasion approximately eightfold compared with the control (Fig. S5b). Therefore, FoxM1 phosphorylation at Ser25 enhanced cell migration and invasion but not cell proliferation.
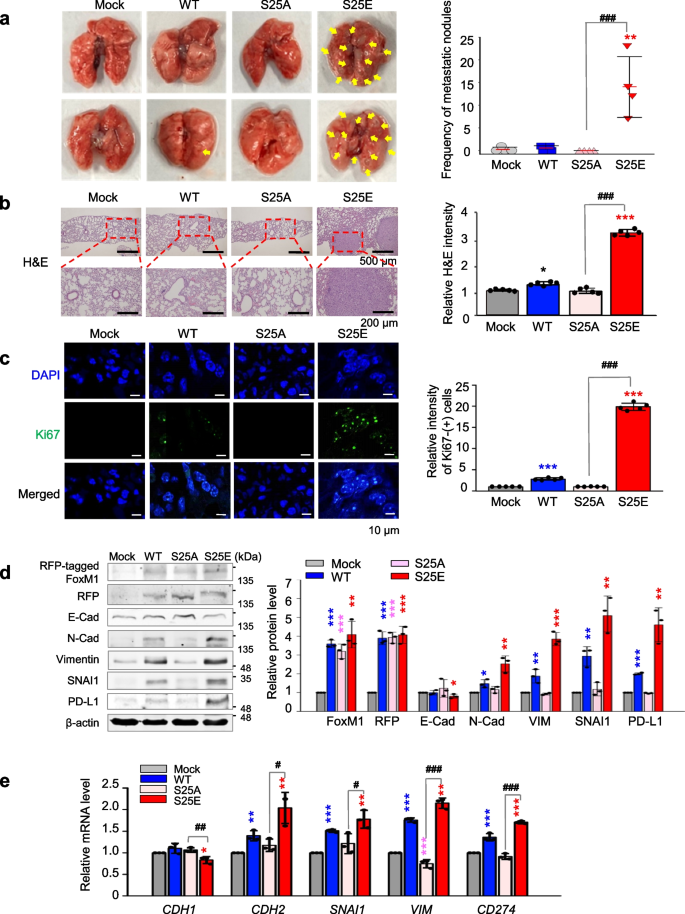
Phosphorylation of FoxM1 at Ser25 facilitates metastasis in a tail-vein injection mouse model
To assess whether phosphorylated FoxM1 at Ser25 triggers metastasis in vivo, A549 cells expressing wild-type (A549WT) or phospho-mutants FoxM1 at Ser25 (A549S25E, A549S25A) were injected into the tail vein of BALB/c nude mice (n ≥ 4) (Fig. S1). In mice injected with A549S25E cells, the frequency of metastatic nodules in the lung was approximately 15 × that of mock (A549Mock) or A549WT (Fig. 4a, right panel). Using mouse lung tissues, H&E staining of cells was higher in tissues injected with A549S25E cells than in those injected with A549Mock, A549WT, or A549S25A (Fig. 4b, right panel). Similarly, Ki67 staining revealed that Ki67-positive cells from mouse lung tissues injected with A549S25E cells were 20 × higher than those of A549Mock or A549S25A (Fig. 4c, right panel). Based on H&E-positive or Ki67-positive cells, lung tissues from mice injected with A549S25E cells showed tumor formation (Fig. 4b-c). Even in mice injected with A549S25A cells, metastatic nodules and proliferation were not found, indicating that phosphorylation of FoxM1 at Ser25 is important in lung metastasis.
Phosphorylation of FoxM1 at Ser25 facilitates metastasis of NSCLC in a tail-vein injection model. A549 cells expressing RFP-tagged WT, S25A, and S25E of FoxM1 were injected intravenously into the tail-veins of four-week-old BALB/c nude mice, and the tumorigenic and metastatic properties were evaluated after 15 weeks. a Representative lung tumor from the mouse model (left panel). The number of metastatic lung tumors was counted and plotted. (n = 4 or 5) (right panel). Data are presented as mean ± SD. b, c Representative H&E staining (b, left panel) and Ki-67 staining (c, left panel) were performed using lung tissue from the mice.The relative density of H&E staining (b, right panel) and Ki-67-positive cells (c, right panel) was analyzed and plotted. *p < 0.05; **p < 0.01; ***p < 0.001. Data are presented as mean ± SD. d Immunoblotting was performed using lung tissue lysates from each mouse model. FoxM1, RFP, E-cadherin, N-cadherin, vimentin, SNAI1, PD-L1, and β-actin were detected using specific antibodies (left panel). The band intensity values were quantified using LI-COR Odyssey software, normalized, and plotted (right panel). e qRT-PCR was performed for CDH1, CDH2, SNAI1, VIM, and CD274 using lung tissue lysates from each mouse model. The relative mRNA expression was plotted. *p < 0.05; **p < 0.01; ***p < 0.001 compared with experimental control. #p < 0.05; ###p < 0.001 compared with S25A FoxM1
Immunoblotting using the lung tissues of mice showed that the levels of N-cadherin, vimentin, and SNAI1 were upregulated in tissues expressing FoxM1S25E compared with the mock system (Fig. 4d). However, E-cadherin level was lower in tissues expressing FoxM1S25E than FoxM1Mock, FoxM1WT, or FoxM1S25A. Consistent with immunoblotting, qRT-PCR analysis showed that the levels of CDH2, SNAI1, and VIM were approximately 2.5 × higher in lung tissues injected with A549S25E than with A549Mock (Fig. 4e), indicating that tissues expressing FoxM1S25E underwent EMT and metastasis. The upregulated mesenchymal factors CDH2, SNAI1, and VIM in A549S25E were downregulated by treatment with thiostrepton, a FoxM1 inhibitor [45] (Fig. S5c). The level of PD-L1 (encoded by CD274) was higher in tissues having A549S25E than A549Mock, A549WT, or A549S25A (Fig. 4d-e). Thus, p-FoxM1Ser25 enhances metastatic lung nodule formation through activation of the EMT and immune escape in an in vivo mouse model.
Interferon signaling is activated in invasive cells with phosphorylated FoxM1
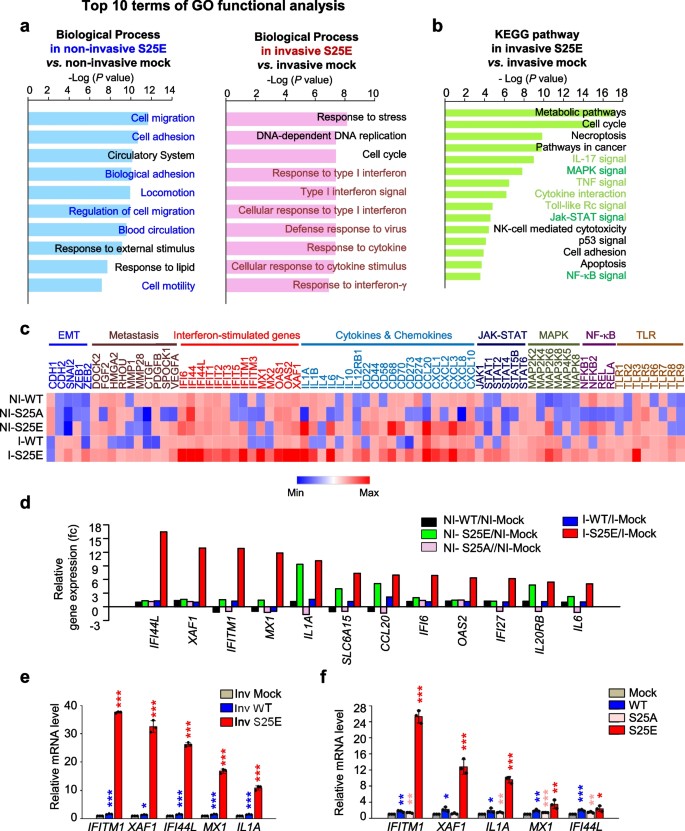
For exploring pathways and factors triggered by p-FoxM1Ser25 depending on invasiveness, microarray analysis was performed. When invasive cells adhered to the external surface of the Transwell chamber filled with Matrigel, non-invasive cells adhered to the inner side of the well. Notably, A549S25A cells were never exhibited invasion. Using invasive and non-invasive cells expressing FoxM1S25E or FoxM1WT, transcriptome profiles (Supplementary Dataset 1) were analyzed with a microarray (Fig. 5). In Gene Ontology (GO) analysis of transcriptome profiles of non-invasive A549S25E cells compared with non-invasive A549Mock cells, genes related to cell migration, adhesion, and the circulatory system were significantly changed in the top three terms of the GO functional analysis, indicating that non-invasive A549S25E cells experienced changes in the levels of genes related to intravasation (Fig. 5a, left panel). After invasion, invasive A549S25E cells showed changes in genes related to interferon signaling and cytokines in the top 10 terms of the GO analysis compared with invasive mock cells (Fig. 5a, right panel). KEGG pathways were analyzed using significantly changed genes having a fold change cutoff ≥ 1.5 (Fig. 5b).Several inflammatory pathways, including IL17, MAPK, TNF, cytokine interaction, Toll-like receptor (TLR), JAK-STAT, and NF-κB signaling, were involved in invasive A549S25E cells relative to A549Mock. The highly expressed genes including interferon-stimulated genes (ISGs) in the microarray were displayed in a heatmap (Fig. 5c). The levels of genes related to EMT, metastasis, cytokines, ISGs, JAK-STAT, MAPK, and NF-κB signaling were highly upregulated in invasive A549S25E cells compared with A549Mock. Inflammatory cytokines such as IL1A, IL1B, and IL6 were highly expressed in invasive A549S25E cells (Fig. 5c), consistent with the increase of cytokine stimuli and responses observed in the biological process (Fig. 5a, right panel) and cytokine interaction detected in the KEGG pathway (Fig. 5b). The relative gene change analysis revealed that IFI44L, XAF1, IFITM1, MX1, and IL1A were the top five most highly expressed of the invasive A549S25E cells among the 53,617 genes analyzed (Fig. 5d). Using qRT-PCR, the expression of IFI44L, XAF1, IFITM1, MX1, and IL1A was reanalyzed in invasive A549 cells (Fig. 5e) and total A549 and HCC827 cells (Fig. 5f, Fig. S5d). IFITM1 was the most highly expressed among invasive cells or total cells expressing FoxM1S25E. IL1A was upregulated in both non-invasive and invasive A549S25E cells (Fig. 5d) and was the third most highly expressed gene in the total A549S25E cells. Therefore, ISGs and IL1A were activated in invasive cells expressing phosphomimetic FoxM1.
Interferon signaling is mainly activated in invasive cells having phosphorylated FoxM1 at Ser25. a, b Using non-invasive and invasive A549 cells expressing S25E of FoxM1, transcriptome profiles were analyzed by microarray. The transcriptome data were clustered by gene probes with fold change > 1.5 in cells expressing S25E FoxM1. Gene Ontology (GO) analysis of transcriptome profiles was performed as biological processes (a) and KEGG pathways (b). KEGG pathways were analyzed, and the signaling pathways had higher gene numbers in cells expressing invasive S25E than in those expressing mock. c Transcriptome comparison between gene profiles of invasive and non-invasive A549 cells expressing WT, S25A, and S25E. The MORPHEUS program was used to visualize the expression levels of genes related to the EMT, metastasis, interferon-stimulated genes, cytokines and chemokines, JAK-STAT, MAPK, NF-kB, and TLR signaling in non-invasive and invasive A549 cells expressing WT, S25A, and S25E of FoxM1. d Relative gene expression profile of the top 12 genes in invasive and non-invasive A549 cells expressing WT, S25E, and S25A FoxM1. e, f qRT-PCR was performed for the top five genes IFITM1, XAF1, IF44L, MX1, and IL1A in invasive A549 cells expressing FoxM1 (e) and total A549 cells expressing FoxM1 (f). *p < 0.05; **p < 0.01; ***p < 0.001; (n = 3). Data are presented as mean ± SD
p-FoxM1S25 triggers cytokine expression for recruitment of macrophages and polarization of M2d-TAM
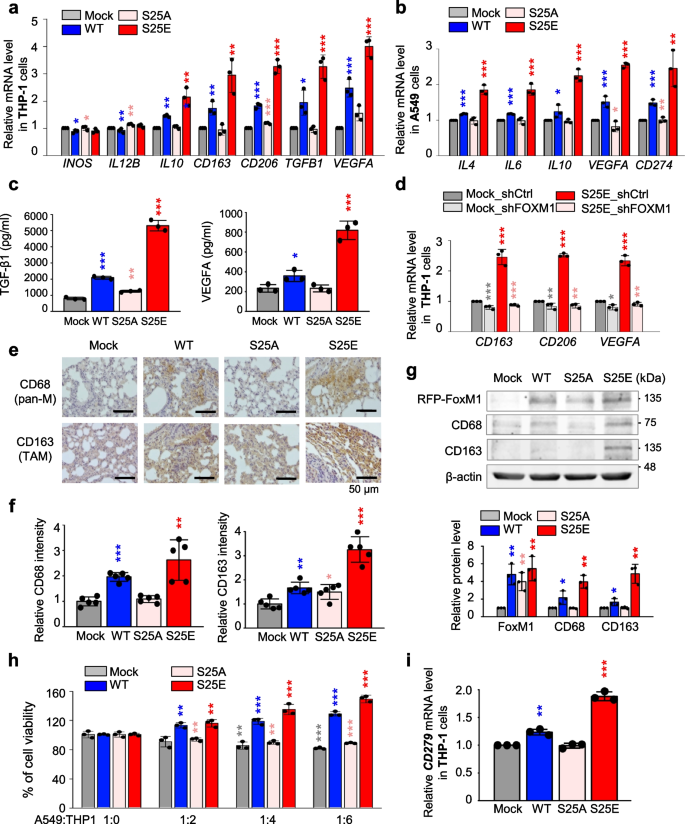
Because FoxM1 is involved in alterations of macrophages [46, 47] and cytokines for macrophages activation are highly expressed in invasive A549S25E cells (Fig. 5), we investigated whether p-FoxM1Ser25 is involved in macrophage polarization of the TME. Expression of the markers of M1 and M2 subtypes was observed in co-cultured human THP-1 and human monocyte-like cells derived from human pluripotent stem cells by qRT-PCR. The levels of M1 markers, INOS and IL12B, did not change significantly in THP-1 cells co-cultured with A549S25E cells (Fig. 6a, Fig. S1). However, the levels of M2 markers IL10, CD163, and CD206 increased up to 2.2, 3.0, and 3.2 times, respectively, in THP-1 cells co-cultured with A549S25E cells compared with A549Mock cells. Because TAMs have characteristics of pro-tumorigenesis and angiogenesis in the TME by release of TGF-β and VEGF [48, 49], the expression of TGFB1 and VEGFA was tested (Fig. 6a). The levels of TGFB1 and VEGFA were 3.2- and 4.0-fold higher, respectively, in THP-1 cells co-cultured with invasive A549S25E cells compared with A549Mock. IL4 and IL10 are universal inducers for M2 type, and IL6 and VEGFA are inducers for M2d [48, 50]. Upregulation of cytokines for inducing M2d subtype, IL4, IL6, IL10, and VEGFA, was observed in A549S25E cells compared with A549Mock (Fig. 6b). This phenomenon was similar in primary human macrophages derived from human pluripotent stem cells co-cultured with invasive A549S25E cells (Fig. S6a). In addition, upregulation of cytokines for inducing M2d subtype, IL4, IL6, IL10, and VEGFA, was observed in A549S25E cells compared with A549Mock cocultured with primary human macrophages (Fig. S6b). Furthermore, the secreted levels of TGF-β1 and VEGFA from the medium of THP-1 cells co-cultured with A549S25E cells (Fig. 6c) or HCC827S25E cells (Fig. S6c), determined by ELISA assay, increased up to 6.6 × and 3.5 × in A549S25E cells, or 5.5 × and 2.0 × in HCC827 S25E cells, respectively, compared with mock cells (Fig. 6c).
p-FoxM1.S25 functions in recruitment of macrophages and triggers polarization of M2-like TAM. a, b Monocyte THP-1 cells were co-cultured with A549 cells expressing mock, WT, S25A, and S25E FoxM1 for 48 h. In THP-1 cells, qRT-PCR was performed for markers of M1 (INOS, IL12B), M2 (IL10, CD163, CD206), and TAM (TGFB1, VEGFA) (a). In A549 cells, qRT-PCR was performed for IL4, IL6, IL10, VEGFA, and CD274 (b). *p < 0.05; **p < 0.01; ***p < 0.001; (n = 3). Data are presented as mean ± SD. c, THP-1 cells were co-cultured with A549 cells expressing mock, WT, S25A, and S25E FoxM1. The secreted levels of TGF-β1 and VEGFA from THP-1 cells co-cultured with A549 cells were detected using ELISA. d Monocyte THP-1 cells were co-cultured with A549 cells expressing mock or S25E depleted FoxM1 using shRNA for 48 h. Using THP-1 cells, qRT-PCR was performed for CD163, CD206, and VEGFA. *p < 0.05; **p < 0.01; ***p < 0.001; (n = 3). e, f Representative CD68 (pan-macrophage marker) staining (e, upper panel) and CD163 (TAM marker) staining (e, lower panel) were performed using lung tissue from mice. The relative density of CD68 staining (f, left panel) and CD163 staining (f, right panel) was analyzed and plotted. *p < 0.05; **p < 0.01; ***p < 0.001. Data are presented as mean ± SD. g Immunoblotting was performed using lung tissue lysates from each mouse model. FoxM1, CD68, CD163 and β-actin were detected using specific antibodies (upper panel). The relative protein intensities were analyzed and plotted (lower panel). h The viability of A549 cells expressing mock, WT, S25A, and S25E FoxM1 was measured when the cells were co-cultured with monocyte THP-1 cells. The ratio between A549 cells and THP-1 cells was 1:0, 1:2, 1:4, and 1:6, as indicated. i Monocyte THP-1 cells were co-cultured with A549 cells expressing mock, WT, S25A, and S25E FoxM1. qRT-PCR was performed for CD279 mRNA level in THP-1 cells. Data are presented as mean ± SD of at least three independent experiments (significantly different from the experimental control). **p < 0.01; ***p < 0.001; (n = 3)
To determine if the presence of FoxM1 is required in TAM differentiation, FoxM1 was depleted using shRNA targeting human FoxM1 (Fig. S6d). After FoxM1-shRNA was treated in A549Mock (A549Mock_shFOXM1) or A549S25E (A549S25E_shFOXM1) (Fig. S6e), THP-1 cells were co-cultured (Fig. 6d). The expression of CD163, CD206, and VEGFA was higher in THP-1 co-cultured with A549S25E_shControl cells than with A549Mock_shControl cells. However, when FoxM1 was depleted, the levels of CD163, CD206, and VEGFA were downregulated in THP-1 cells co-cultured with A549S25E_shFOXM1 cells compared with the control (Fig. 6d). Therefore, upregulation of cytokines such as IL4, IL6, and IL10 in invasive A549S25E cells facilitated polarization of the M2d subtype of macrophage, which was eliminated by depletion of FoxM1 in the cocultured system.
Then, to investigate whether macrophages were recruited in TME by A549S25E cells showing high expression of VEGFA [50] and IL1A [51], mouse tumor tissues were stained with specific macrophage antibodies for TAM marker CD163 and pan-macrophage marker CD68. Consistently, the relative CD68-positive intensity was ~ 2.6 × higher in lung tissues from mice injected with A549S25E cells than with A549Mock cells (Fig. 6e-f). The relative intensity of CD163-positive cells was 3.2 × higher in lung tissues from mice injected with A549S25E cells than with A549Mock cells (Fig. 6f). Immunoblotting using tissues of mice injected with A549S25E cells revealed that CD163 and CD68 were highly expressed in TME of A549S25E (Fig. 6g). Accordingly, macrophages were recruited and TAMs were induced in the TME of lung tissues from mice injected with LUAD expressing FoxM1S25E. We also investigated whether inhibition of TGF-β signaling reduced the levels of mesenchymal markers and IL6. Treatment of TGF-β-specific inhibitor SB431542 reduced the levels of p-PLK1, FoxM1, IFITM1, p-Smad2/3, IL6, IL1A, N-cadherin, SNAI1, and vimentin (Fig. S7), indicating that TGF-β signaling is important to FoxM1 phosphorylation-mediated downstream events including EMT and cytokine secretion for TAM polarization (Fig. S7).
PD-1 expression by TAMs regulates tumor immunity [52]. The higher level of CD274 (encoding PD-L1, a ligand of PD-1) in mouse lung tissues injected with A549S25E or in A549S25E cells co-cultured with THP-1 or primary human macrophage cells was higher than that with A549Mock, A549WT, or A549S25A cells (Fig. 4e, Fig. 6b, Fig. S6b, Fig. S6f), indicating viability of A549S25E cells co-cultured with macrophages. The viability of A549S25E cells increased at a ratio of 1:6 (A549:THP-1 cells) compared with A549Mock cells (Fig. 6h). The increase in LUAD cell viability may be attributed to the potential interaction between PD-L1 of A549S25E and PD-1 of THP-1 cells based on the previous studies [53] (Fig. 6h-i). Therefore, PD-L1 upregulation induced by FoxM1S25E would evade the immune checkpoint by polarization of M2d-TAM that expressed PD-1 in the TME.
p-FoxM1S25 directly activates the expression of IL1, IL6, VEGFA, SNAI1, and PD-L1 but not IFITM1 that are regulated by STING/TBK1/IRF3, JAK1/STAT1, and/or MEK signaling
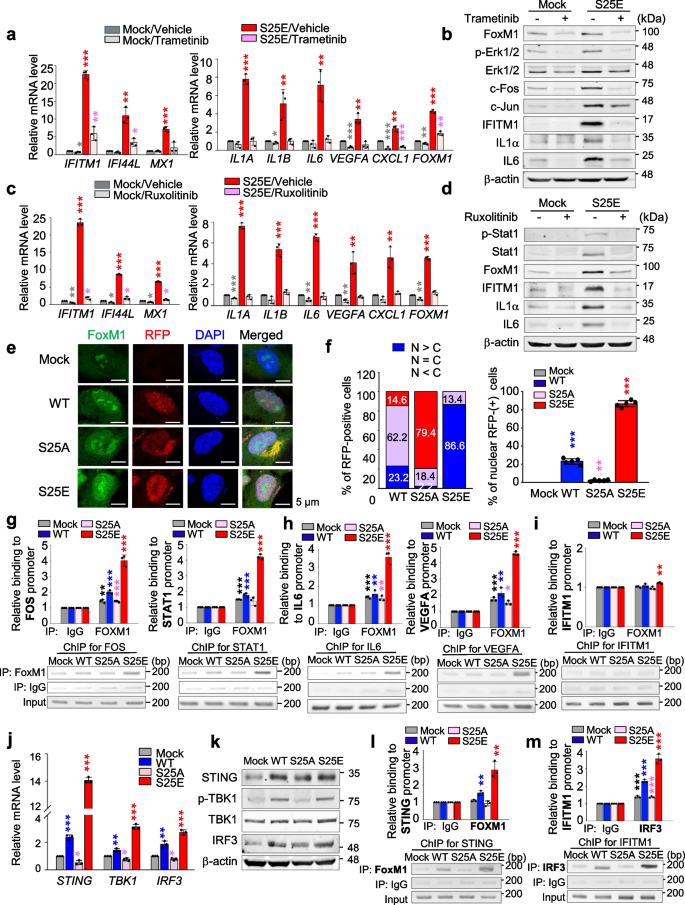
ISGs including IFITM1, IFI44L, and MX1 were highly expressed in invasive A549S25E cells compared with the control (Fig. 5). ISGs were induced by several signaling pathways, including MAPK, JAK-STAT, and NF-κB signaling [54], which were detected in GO analysis (Fig. 5b) and APPYTER analysis (Fig. S8a-b) of the microarray data of invasive A549S25E. To elucidate the signaling pathway for expression of IFITM1 in invasive A549S25E cells, trametinib, ruxolitinib, fludarabine, and BI605906 were applied for inhibiting MEK, JAK1, STAT1, and IKKβ, respectively (Fig. 7a-d, Fig. S9a-d). The levels of IFITM1, IFI44L, and MX1 were markedly downregulated by treatment with trametinib, ruxolitinib, and fludarabine but not by BI605906 in A549S25E cells, while treatment with trametinib, ruxolitinib, fludarabine, and BI605906 markedly reduced the mRNA levels of IL1A, IL1B, VEGFA, and IL6 for macrophage recruitment or TAM polarization [51] (Fig. 7a and c, Fig. S9a and S9c). Moreover, the levels of CDH2, VIM, and SNAI1/2 were downregulated by treatment with trametinib, ruxolitinib, and fludarabine, but not by treatment with BI605906 (Fig. S9e-h). Thus, ISGs and mesenchymal factors upregulated in A549S25E can be activated by MEK and JAK1/STAT1 pathways but not by NF-kB.
p-FoxM1S25 translocates to the nucleus and activates genes for monocyte recruitment, TAM polarization, immune escape, and angiogenesis by direct activation. a, b A549S25E were treated with 1 μM trametinib, an inhibitor of MEK, for 48 h. a qRT-PCR was performed for interferon-stimulated genes (IFITM1, IF44L, and MX1), IL1A, IL1B, IL6, VEGFA, CXCL1, and FOXM1 in A549S25E cells. b Immunoblot analyses were performed using specific antibodies for FoxM1, p-Erk1/2, Erk1/2, c-Fos, c-Jun, IFITM1, IL1A, IL6, and β-actin. c, d A549S25E cells were treated with 15 μM ruxolitinib, a JAK inhibitor, for 48 h. c qRT-PCR was performed for IFITM1, IF44L, MX1, IL1A, IL1B, IL6, VEGFA, CXCL1, and FOXM1 in A549S25E cells. d Immunoblot analyses were performed using anti-FoxM1, anti-STAT1, anti-p-STAT1, anti-IFITM1, anti-IL1A, anti-IL6, and anti-β-actin. e Immunofluorescence was performed with A549 cells expressing WT, S25A, or S25E mutant of FoxM1. FoxM1 (green), RFP (red), and DNA (DAPI, blue) was displayed. Scale bar, 5 μm. f The quantification of the population of cells in the cytoplasm, nucleus, and both is presented on the left. The percentage of cells that exhibited positive RFP (red) staining was assessed with the following categories (N > C, RFP staining predominantly in the nucleus; N = C, similar RFF levels in both the nucleus and cytoplasm; N < C, RFP staining mainly in the cytoplasm). The population of RFP-positive cells specifically in the nucleus was plotted (right). n > 800. g-i ChIP assays for FoxM1 binding to the promoters of FOS (g, left), STAT1 (g, right), IL6 (h, left), VEGFA (h, right), and IFITM1 (i). Assays were performed on chromatin fragments using antibody to FoxM1 and normalized to pre-immune normal IgG. Immunoprecipitated fractions were assayed by qRT-PCR for binding the promoters of FOS, STAT1, IL6, VEGFA, and IFITM1. The qRT-PCR products were visualized in agarose-gel. j qRT-PCR was performed for STING, TBK1, and IRF3 in A549S25E cells. k Immunoblot analyses were performed using specific antibodies for STING, p-TBK1, TBK1, IRF3 and anti-β-actin. l ChIP assays were performed for FoxM1 binding to the promoters of STING in A549S25E cells. m ChIP assays for IRF3 binding to the promoters of IFITM1 in A549.S25E cells. Assays were performed on chromatin fragments using antibody to IRF3 and normalized to pre-immune normal IgG. Immunoprecipitated fractions were assayed by qRT-PCR for binding the promoters of IFITM1. Data are presented as mean ± SD of three independent experiments (significantly different from the experimental control). *p < 0.05; **p < 0.01; ***p < 0.001; (n = 3)
For further investigation of whether p-FoxM1S25 upregulates its transcriptional activity, the nuclear location of FoxM1S25E was observed by immunostaining (Fig. 7e-f, Fig. S10). Up to 86.6% of RFP-tagged FoxM1S25E was located in the nucleus, while only 2.2% of FoxM1S25A was located in the nucleus (Fig. 7e-f, Fig. S10), indicating that FoxM1S25E dominantly translocates into the nucleus. To determine whether p-FoxM1S25 can directly bind the promoter regions of genes for proinflammation, EMT, immune escape, or ISGs, conserved binding sequences around promoter regions of these genes were analyzed (Fig. S11a). A ChIP assay using anti-FoxM1 antibody revealed that FoxM1S25E directly bound the promoters of FOS, STAT1, IL6, VEGFA, IL1A, IL1B, SNAI1, and CD274 and upregulated their expression by 3–4 × compared with mock (Fig. 7g-h, Fig. S11b-e). However, FoxM1 did not bind to the promoter of IFITM1 (Fig. 7i). Thus, p-FoxM1S25 directly activates the expression of genes for monocyte recruitment, TAM polarization, angiogenesis, immune escape, and EMT.
Next, we wanted to investigate which factors regulate the expression of IFITM1. Because STING-TBK1-IRF3 signaling is a well-established upstream regulator of IFN genes to mediate immune defense [55], the levels of STING, TBK1, and IRF3 was observed in A549S25E cells. qRT-PCR analysis revealed that the expressions of STING, TBK1, and IRF3 were highly upregulated in A549S25E cells compared with A549Mock cells (Fig. 7j). In addition, the levels of active p-TBK1 increased in A549S25E cells compared with A549Mock cells by immunoblot analysis (Fig. 7k). A ChIP assay using anti-FoxM1 antibody revealed that FoxM1S25E directly bound the promoter of STING and upregulated the expression of STING by 3 × in A549S25E cells compared with A549Mock (Fig. 7l, Fig. S11f). Additional ChIP analysis using anti-IRF3 showed that IRF3 bound to the promoter regions of IFITM1 and upregulated the expression of IFITM1 by 4 × in A549S25E cells compared with A549Mock cells (Fig. 7m, Fig. S11g), indicating that FoxM1S25E upregulates the expression of STING, which triggers the expression of TBK1 and IRF3. Consequently, IRF3 activates the expression of IFITM1 in cells expressing FoxM1S25E. Therefore, the expression of IFITM1 is regulated through the STING-TBK1-IRF3 pathway that is activated by p-FoxM1S25.
IFITM1 induces invasiveness and TAM polarization in invasive FoxM1- or TGF-β-induced EMT
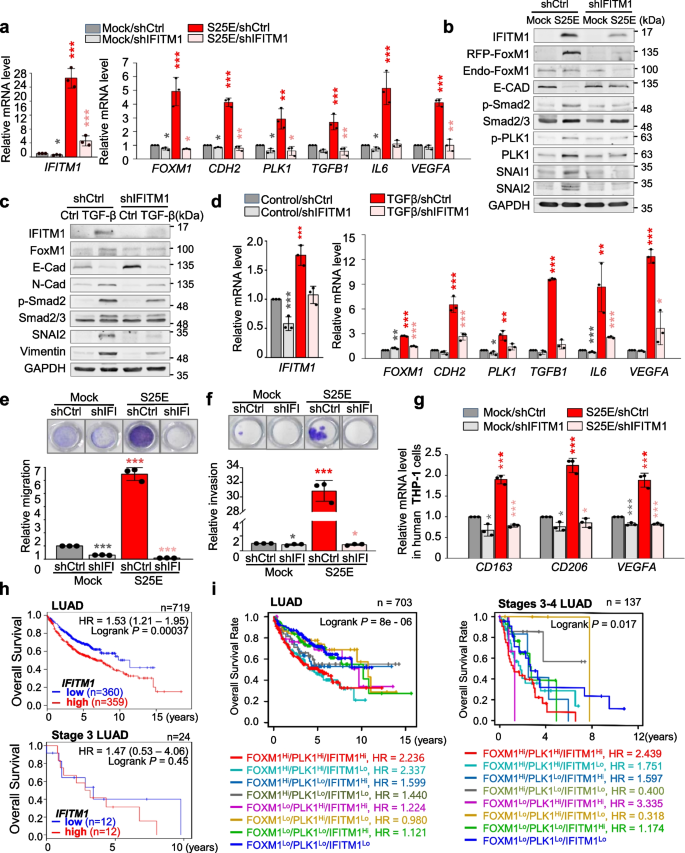
In A549S25E cells, IFITM1 was the highest expressed factor among the top five genes (Fig. 5e-f). To determine the effect of IFITM1 on metastasis and macrophage polarization, IFITM1 was depleted with viral shRNA targeting human IFITM1 (Fig. 8, Fig. S12a). When IFITM1 expression was downregulated in IFITM1-depleted A549S25E cells, the levels of CDH2, TGFB1, IL6, and VEGFA upregulated by FoxM1S25E were markedly reduced compared with the scramble control (Fig. 8a), indicating IFITM1 affects p-FoxM1S25-mediated EMT activation and TAM polarization. In addition, the levels of IL6 and VEGFA were reduced by IFITM1 knock-down in A549S25E (A549S25E_shIFITM1) cells compared with those of A549S25E cells treated with scramble shRNA control (A549S25E_shControl). Immunoblot analysis revealed that levels of IFITM1, FoxM1, p-Smad2, and SNAI1 were lower in A549S25E_shIFITM1 than A549S25E_shControl (Fig. 8b). Notably, p-PLK1T210 level was also downregulated in A549S25E_shIFITM1 compared with that of A549S25E_shControl. FoxM1 or IFITM1 depletion mutually reduced each level (Fig. 8a, Fig. S12b), while exogenous expression of FoxM1 or IFITM1 mutually upregulated each other in A549 cells (Fig. 5, Fig. S12c-d), suggesting that IFITM1 and FoxM1 closely but not directly regulate mutual expression (Fig. 7i). When IFITM1 was depleted during TGF-β-induced EMT, the upregulation of N-cadherin, vimentin, SNAI2, and p-Smad2 by TGF-β treatment was reduced by IFITM1 knock-down (Fig. 8c). Consistent with Fig. 8a, IFITM1 depletion suppressed the expression of CDH2, TGFB1, IL6, and VEGFA more strongly than that of control shRNA in TGF-β-induced EMT (Fig. 8d). Because IFITM1 depletion reduced the expression of mesenchymal markers and TGFB1 (Fig. 8a-d), the motility and invasiveness were determined using Transwell and/or the invasion system. Motility and invasiveness in A549S25E_shIFITM1 were reduced approximately 6- and 30-fold, respectively, compared with A549S25E_shControl cells (Fig. 8e-f), indicating that IFITM1 is important in the progression of cell migration and invasion. To further investigate whether knock-down of IFITM1 can affect M2-like TAM polarization, M2d markers CD163, CD206, and VEGFA were observed in THP-1 cells co-cultured with A549S25E_shIFITM1 (Fig. 8g). The M2d markers were more markedly reduced in THP-1 cells co-cultivated with A549S25E_shIFITM1 cells than in A549S25E_shControl cells (Fig. 8g), indicating that IFITM1 is an important trigger of M2d-TAM polarization. Therefore, IFITM1 upregulated by p-FoxM1S25 should be important to regulate cancer metastasis and TAM polarization in the TME.
IFITM1 functions as a regulator of metastasis and polarization of M2d-TAM in p-FoxM1-induced metastasis. a, b IFITM1 was depleted in A549S25E cells using human IFITM1 shRNA for 48 h. a qRT-PCR was performed for IFITM1, FOXM1, CDH2, PLK1, TGFB1, IL6, and VEGFA. b Immunoblot analyses were performed using anti-IFITM1, anti-RFP, anti-E-cadherin, anti-p-Smad2, anti-Smad2/3, anti-SNAI1, anti-SNAI2, and anti-GAPDH antibodies. c, d 5 ng/mL TGF-β was applied to A549 cells depleting IFITM1 with shRNA. c Immunoblotting was performed using anti-IFITM1, anti-FoxM1, anti-E-cadherin, anti-N-cadherin, anti-p-Smad2, anti-Smad2/3, anti-SNAI2, anti-vimentin, and anti-GAPDH antibodies. d qRT-PCR was performed for IFITM1, FOXM1, CDH2, PLK1, TGFB1, IL6, and VEGFA. e IFITM1 was depleted in A549S25E cells using human IFITM1 shRNA for 48 h. Cells were subjected to a Transwell migration assay. Three days after seeding, the cells on the bottom surface were stained with 0.05% crystal violet dye. Images of the Transwell cell migration assay were collected and analyzed with an Odyssey infrared imaging system and plotted. f An invasion assay was performed using A549 cells expressing wild-type or mutants of FoxM1. Seven days after seeding, the cells that invaded the bottom surface were stained with 0.05% crystal violet dye, and the relative absorbance was plotted (n = 3). g THP-1 cells were co-cultured with A549S25E cells depleting IFITM1 for 48 h. qRT-PCR was performed for CD163, CD206, and VEGFA. h The overall survival (OS) of all LUAD patients (n = 719) (h, upper) and stage 3 LUAD patients (n = 24) (h, lower) was analyzed according to IFITM1 expression level using KM PLOTTER. High (Hi) and low (Lo) were generated based on the expression at the median cut-off. i The OS of all LUAD patients (n = 703) (i, left) and stage 3–4 LUAD patients (n = 137) (i, right) was analyzed according to IFITM1, FOXM1, and PLK1 expression levels using KM PLOTTER. High (Hi) and low (Lo) were generated based on the expression at the median cut-off. Data are presented as mean ± SD of three independent experiments (significantly different from the experimental control). *p < 0.05; ***p < 0.001; (n = 3)
Clinical relevance of PLK1, IFITM1, and FOXM1 in advanced LUAD
We next investigated the clinical relevance between IFITM1 expression and survival rates of advanced LUAD in patients. The analysis (Fig. 8h, Table S9) showed that IFITM1 expression is significantly correlated with OS of LUAD patients (Fig. 8h, upper panel; n = 719, HR = 1.53, log rank P = 0.00037) but not that of LUSQ patients (Fig. S12e; n = 524, HR = 0.92, log rank P = n.s.). Additional analysis between OS and co-expression of IFITM1, FOXM1, and PLK1 in LUAD patients showed that the OS with high FOXM1/PLK1/IFITM1 expression was significantly shorter than that with low FOXM1/PLK1/IFITM1 expression (n = 703, log-rank P = 8e-06) (Fig. 8i, left panel). Furthermore, in a clinical analysis of advanced LUAD patients at stages 3–4, the OS of patients with high FOXM1/PLK1/IFITM1 expression was shorter than for those with low FOXM1/PLK1/IFITM1 (n = 137, log-rank P = 0.017) (Fig. 8i, right panel). The cumulative OS in LUAD patients revealed that high expression of at least two of PLK1, IFITM1, and FOXM1 highly correlated with OS of advanced LUAD patients as well as all stages of LUAD (Fig. 8i). To understand IFITM1 expression in metastatic LUAD, a heatmap was analyzed based on the degree of expression of IFITM1 from TCGA data analysis in normal and tumor tissue, depending on stage of LUAD (Fig. S12f). Concurrent expression of IFITM1, FOXM1, and PLK1 showed a high correlation with tumor formation in primary and advanced LUAD but not in LUSQ. Therefore, additional expression of IFITM1 with FOXM1 and PLK1 should affect the survival rates of advanced LUAD.





Add Comment