Exposure to DEP results in increased ACE2 protein expression in the bronchioles of C57BL/6 mice
To investigate whether subchronic inhalational exposure to DEP and consumption of a HF diet alter ACE2 expression, we measured ACE2 protein expression by immunofluorescent histology (Fig. 1A-M) and mRNA transcript expression by RT-qPCR (Fig. 1N). We observed a significant increase in ACE2 protein expression in experimental groups that were exposed to DEP, regardless of diet. Compared with that in the LF + CON group, ACE2 protein expression was significantly greater in the LF + DEP (p < 0.0001) and HF + DEP groups (p < 0.0001). Compared with that in the HF + CON group, there was a significant increase in ACE2 protein expression in the LF + DEP (p < 0.0001) and HF + DEP groups (p < 0.0001). There was no significant difference in protein expression observed among the exposure groups. Exposure F = 71.07, diet F = 2.494, and exposure x diet F = 0.08277. There was no significant difference in ACE2 mRNA transcript expression between the groups.
Exposure to DEP results in increased angiotensin-converting enzyme 2 (ACE2) receptor protein expression in the bronchioles of C57BL/6 mice. Representative images of ACE2 expression in the bronchioles of C57Bl/6 mice on a control (LF; A–C) or high-fat (HF; G–I) diet exposed to saline (CON) or on a LF (D–F) or HF diet (J–L) exposed to diesel exhaust particles (DEP – 35 µg PM) twice a week for a total of 30 days. Red fluorescence indicates ACE2 expression, and blue fluorescence indicates nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung ACE2 mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of ACE2 mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification; scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to LF + CON, †p < 0.05 compared to LF + DEP, ‡p < 0.05 compared to HF + CON by two-way ANOVA. The brightness of the nuclei image panels was increased by 40% and the overlay images were increased by 10%, as shown in this figure
Probiotic treatment mitigated the DEP-mediated increase in ACE2 receptor protein expression in the lungs of C57BL/6 mice
To determine whether probiotic treatment mitigates DEP- ± HF diet-mediated alterations in ACE2 expression, we measured ACE2 protein expression via immunofluorescent histology (Fig. 2A-M) and mRNA transcript expression by RT-qPCR (Fig. 2N). We observed a significant decrease in ACE2 protein expression in the HF + CON + PRO (p < 0.0001) and HF + DEP + PRO (p < 0.0001) groups compared with the HF + DEP group. There was no significant difference in ACE2 protein expression between the probiotic groups and the HF + CON group. Exposure F = 43.91, probiotics F = 38.18, exposure x probiotics F = 46.77.
We also observed a significant decrease in ACE2 mRNA expression in the HF + CON + PRO (p = 0.0012) and HF + DEP + PRO (p = 0.0017) groups compared to the HF + CON group. Similarly, ACE2 mRNA expression was significantly lower in the HF + CON + PRO (p = 0.0003) and HF + DEP + PRO (p = 0.0005) groups than in the HF + DEP group. There was no significant difference in ACE2 mRNA expression between the HF + CON and HF + DEP groups or between the probiotic treated groups. Exposure F = 32.10, probiotics F = 0.5418, exposure x probiotics F = 0.4563.
Probiotic treatment mitigates the DEP-mediated increase of angiotensin-converting enzyme 2 (ACE2) protein expression in the bronchioles of C57BL/6 mice. Representative images of ACE2 expression in the bronchioles of C57Bl/6 mice on a high-fat (HF) diet exposed to either (A–C) saline (CON), (D–F) DEP – 35 µg PM, (G–I) saline and probiotics (PRO) − 0.3 g/day (~ 7.5 × 107 cfu/day), or (J–L) DEP and probiotics. Red fluorescence indicates ACE2 expression, the blue fluorescence indicates nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung ACE2 mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of ACE2 mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification, scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to HF + CON, †p < 0.05 compared to HF + DEP, ‡p < 0.05 compared to HF + CON + PRO by two-way ANOVA. The brightness of the nuclei image panels was increased by 40% and the overlay images were increased by 10%, as shown in this figure
Exposure to diesel exhaust particulate matter in conjunction with consumption of a HF diet results in increased TMPRSS2 expression in the lungs of C57BL/6 mice
To assess whether subchronic inhalational exposure to DEP and consumption of a HF diet alter TMPRSS2 expression, we measured TMPRSS2 protein expression by immunofluorescent histology (Fig. 3A-M) and mRNA transcript expression by RT-qPCR (Fig. 3N). We observed a significant increase in TMPRSS2 protein expression in the HF + DEP group compared to the LF + CON (p = 0.0004), LF + DEP (p < 0.0001), and HF + CON (p < 0.0001) groups. Compared with the LF + CON group, the LF + DEP (p < 0.0001) and HF + CON (p = 0.0004) groups presented significantly lowerTMPRSS2 protein expression. Compared to the LF + DEP group, the HF + CON (p = 0.0273) group displayed significantly increased TMPRSS2 protein expression was significantly increased. Exposure F = 1.325, diet F = 16.98, exposure x diet F = 56.07.
TMPRSS2 mRNA expression was not significantly different between the LF + CON, LF + DEP, and HF + CON groups, but the expression of TMPRSS2 mRNA in the HF + DEP group was significantly greater than that in the LF + DEP (p = 0.0016) and HF + CON (p = 0.0067) groups. Exposure F = 0.7838, diet F = 2.562, exposure x diet F = 12.77.
Exposure to DEP in conjunction with consumption of a HF diet results in increased transmembrane serine protease 2 (TMPRSS2) expression in the bronchioles of C57BL/6 mice. Representative images of TMPRSS2 expression in the bronchioles of C57Bl/6 mice on a control (LF; A–C) or high-fat (HF; G–I) diet exposed to saline (CON) or fed a LF (D–F) or HF diet (J–L) and exposed to diesel exhaust particles (DEP – 35 µg PM) twice a week for a total of 30 days. Red fluorescence indicates TMPRSS2 expression, and blue fluorescence indicates nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung TMPRSS2 mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of TMPRSS2 mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification; scale bar = 100 μm. The data are presented as mean ± SEM, with *p < 0.05 compared to LF + CON, †p < 0.05 compared to LF + DEP, ‡p < 0.05 compared with HF + CON by two-way ANOVA
Probiotic treatment mitigated the DEP- and HF diet-mediated increase of TMPRSS2 protein expression in the lungs of C57BL/6 mice
To establish whether probiotic treatment mitigates DEP- ± HF diet-mediated alterations in TMPRSS2 expression, we measured TMPRSS2 expression by immunofluorescent histology (Fig. 4A-M) and mRNA transcript expression by RT-qPCR (Fig. 4N). We observed a significant decrease in TMPRSS2 protein expression in groups treated with probiotics (HF + CON + PRO (< 0.0001), HF + DEP + PRO (< 0.0001)) compared to the HF + DEP group. Within the probiotic groups, TMPRSS2 protein expression was observed to be significantly decreased in the HF + DEP + PRO (p = 0.0049) group compared to the HF + CON + PRO group. Exposure F = 20.93, probiotics F = 3.547, exposure x probiotics F = 38.84.
TMPRSS2 mRNA expression was not found to be statistically different between the HF + CON, HF + DEP, and HF + DEP + PRO groups. TMPRSS2 mRNA expression in the HF + CON + PRO (p = 0.0275) group was observed to be significantly lower than that of the HF + DEP group. Exposure F = 2.137, probiotics F = 4.328, exposure x probiotics F = 0.1592.
Probiotic treatment mitigated the DEP- and HF diet-mediated increase in transmembrane serine protease 2 (TMPRSS2) protein expression in the bronchioles of C57BL/6 mice. Representative images of TMPRSS2 expression in the bronchioles of C57Bl/6 mice fed a high-fat (HF) diet exposed to either (A-C) saline (CON), (D-F) DEP – 35 µg PM, (G-I) saline and probiotics (PRO) − 0.3 g/day (~ 7.5 × 107 cfu/day), or (J-L) DEP and probiotics. Red fluorescence indicates TMPRSS2 expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung TMPRSS2 mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of TMPRSS2 mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification, scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to HF + CON, †p < 0.05 compared to HF + DEP, ‡p < 0.05 compared to HF + CON + PRO by two-way ANOVA
Exposure to diesel exhaust particulate matter in conjunction with the consumption of a HF diet results in increased androgen receptor protein expression in the bronchioles of C57BL/6 mice
To determine if subchronic inhalational exposure to DEP and consumption of a HF diet alters AR expression, we measured AR protein expression by immunofluorescent histology (Fig. 5A-M) and mRNA transcript expression by RT-qPCR (Fig. 5N). We observed a significant increase in AR protein expression in the HF + DEP group compared with the LF + CON (p < 0.0001), LF + DEP (p < 0.0001), and HF + CON (p < 0.0001) groups. There was no significant difference observed between the LF + CON, LF + DEP, and HF + CON groups. Exposure F = 10.21, diet F = 17.17, exposure x diet F = 22.58.
AR mRNA expression was significantly greater in the LF + DEP (p = 0.0188) group than in the LF + CON group as well as decreased in the HF + DEP group (p = 0.0307) compared to the LF + DEP group. Exposure F = 1.669, diet F = 1.270, exposure x diet F = 5.069.
Exposure to diesel exhaust particulate matter in conjunction with consumption of a HF diet results in increased androgen receptor (AR) protein expression in the bronchioles of C57BL/6 mice. Representative images of AR expression in the bronchioles of C57Bl/6 mice on a control (LF; A-C) or high-fat (HF; G-I) diet exposed to saline (CON) or on a LF (D-F) or HF diet (J-L) exposed to diesel exhaust particles (DEP – 35 µg PM) twice a week for a total of 30 days. Red fluorescence indicates AR expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung AR mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of AR mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification; scale bar = 100 μm. *p < 0.05 compared to LF + CON, †p < 0.05 compared to LF + DEP, ‡p < 0.05 compared to HF + CON by two-way ANOVA
Probiotic treatment mitigated the DEP- and HF diet-mediated increase in androgen receptor protein expression in the bronchioles of C57BL/6 mice
To ascertain whether probiotic treatment mitigates DEP- ± HF diet-mediated alterations in AR expression, we measured AR expression via immunofluorescent histology (Fig. 6A-M) and mRNA transcript expression by RT-qPCR (Fig. 6N). We observed a significant decrease in AR protein expression in the groups treated with probiotics [HF + CON + PRO (p < 0.0001), HF + DEP + PRO (p < 0.0001)] compared with the HF + DEP group. There was no significant difference observed between the HF + CON and probiotic groups. Exposure F = 14.03, probiotics F = 5.946, exposure x probiotics F = 28.60. There was no significant difference in AR mRNA expression observed between groups.
Probiotic treatment mitigated the DEP- and HF diet-mediated increase of androgen receptor (AR) protein expression in the bronchioles of C57BL/6 mice. Representative images of AR expression in the bronchioles of C57Bl/6 mice on a high-fat (HF) diet exposed to either (A-C) saline (CON), (D-F) DEP – 35 µg PM, (G-I) saline and probiotics (PRO) − 0.3 g/day (~ 7.5 × 107 cfu/day), or (J-L) DEP and probiotics. Red fluorescence indicates AR expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung AR mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of AR mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification, scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to HF + CON, †p < 0.05 compared to HF + DEP, ‡p < 0.05 compared to HF + CON + PRO by two-way ANOVA
Exposure to diesel exhaust particulate matter in conjunction with the consumption of a HF diet results in increased NRP1 protein expression in the bronchioles of C57BL/6 mice
To investigate whether subchronic inhalational exposure to DEP and consumption of a HF diet alter NRP1 expression, we measured NRP1 protein expression via immunofluorescent histology (Fig. 7A-M) and mRNA transcript expression by RT-qPCR (Fig. 7N). We observed a significant increase in NRP1 protein expression in the HF + DEP group compared with the LF + CON (p < 0.0001), LF + DEP (p < 0.0001), and HF + CON (p < 0.0001) groups. There was no significant difference observed between the two LF groups. NRP1 protein expression was decreased in the HF + CON group compared with the two LF groups [LF + CON (p = 0.0159), LF + DEP (p < 0.0001)]. Exposure F = 44.21, diet F = 2.039, exposure x diet F = 27.47.
NRP1 mRNA expression was observed to be significantly greater in the LF + DEP group than in the LF + CON (p = 0.0205), HF + CON (p = 0.0125), and HF + DEP (p = 0.0239) groups. There was no significant difference in NRP1 mRNA expression among the LF + CON, HF + CON, and HF + DEP groups. Exposure F = 3.732, diet F = 3.976, exposure x diet F = 2.236.
Exposure to diesel exhaust particulate matter in conjunction with consumption of a HF diet results in increased neuropilin 1 (NRP1) protein expression in the bronchioles of C57BL/6 mice. Representative images of NRP1 expression in the bronchioles of C57Bl/6 mice on a control (LF; A-C) or high-fat (HF; G-I) diet exposed to saline (CON) or on a LF (D-F) or HF diet (J-L) exposed to diesel exhaust particles (DEP – 35 µg PM) twice a week for a total of 30 days. Red fluorescence indicates NRP1 expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung NRP1 mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of NRP1 mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification; scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to LF + CON, †p < 0.05 compared to LF + DEP, ‡p < 0.05 compared to HF + CON by two-way ANOVA
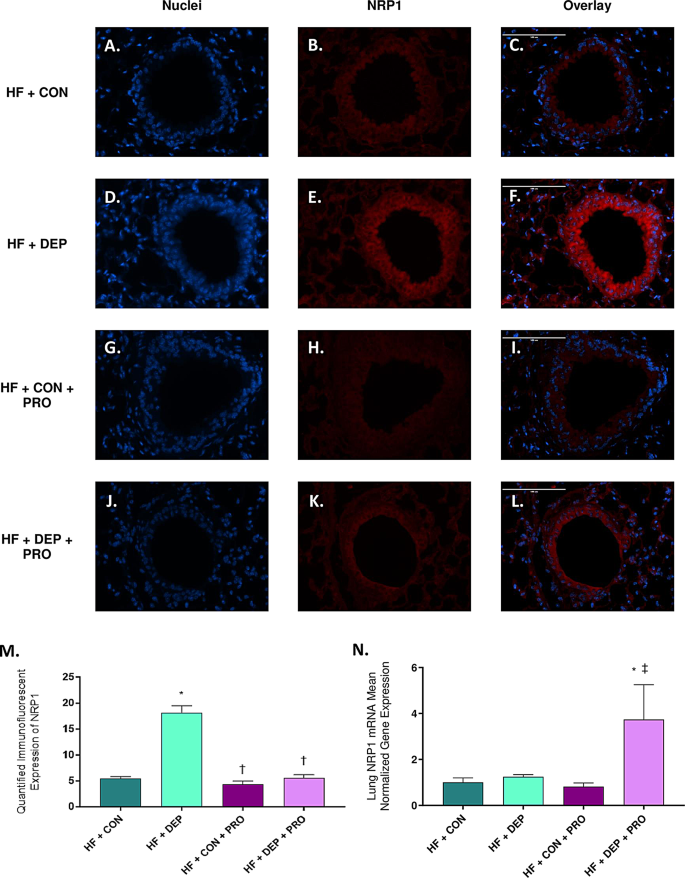
Probiotic treatment mitigated the DEP- and HF diet-mediated increase in NRP1 protein expression in the bronchioles of C57BL/6 mice
To determine whether probiotic treatment mitigates DEP- ± HF diet-mediated alterations in NRP1 expression, we measured NRP1 expression via immunofluorescent histology (Fig. 8A-M) and mRNA transcript expression by RT-qPCR (Fig. 8N). We observed a significant decrease in NRP1 protein expression in groups treated with probiotics [HF + CON + PRO (p < 0.0001), HF + DEP + PRO (p < 0.0001)] compared with the HF + DEP group. There was no significant difference observed between the HF + CON and probiotic groups. Exposure F = 38.85, probiotics F = 41.20, exposure x probiotics F = 27.18.
We also observed a significant increase in NRP1 mRNA expression in the HF + DEP + PRO group compared with the HF + CON (p = 0.0345) and HF + CON + PRO (p = 0.0161) groups. There was no significant difference in NRP1 mRNA expression among the HF + CON, HF + DEP, and HF + CON + PRO groups. Exposure F = 1.792, probiotics F = 3.385, exposure x probiotics F = 2.405.
Probiotic treatment mitigates DEP- and HF diet-mediated increase of neuropilin 1 (NRP1) protein expression in the bronchioles of C57BL/6 mice. Representative images of NRP1 expression in the bronchioles of C57Bl/6 mice on a high-fat (HF) diet exposed to either (A-C) saline (CON), (D-F) DEP – 35 µg PM, (G-I) saline and probiotics (PRO) − 0.3 g/day (~ 7.5 × 107 cfu/day), or (J-L) DEP and probiotics. Red fluorescence indicates NRP1 expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung NRP1 mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of NRP1 mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification, scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to HF + CON, †p < 0.05 compared to HF + DEP, ‡p < 0.05 compared to HF + CON + PRO by two-way ANOVA
Exposure to diesel exhaust particulate matter in conjunction with consumption of a HF diet results in increased furin protein expression in the bronchioles of C57BL/6 mice
To establish whether subchronic inhalational exposure to DEP and consumption of a HF diet alter furin expression, we measured furin expression via immunofluorescent histology (Fig. 9A-M) and mRNA transcript expression by RT-qPCR (Fig. 9N). We observed a significant increase in furin protein expression in the HF + DEP group compared with the LF + CON (p < 0.0001), LF + DEP (p < 0.0001), and HF + CON (p < 0.0001) groups. There was no significant difference observed between the LF + CON, LF + DEP, and HF + CON groups. Exposure F = 30.95, diet F = 28.62, and exposure x diet F = 45.02. We also observed no significant difference in furin mRNA expression among the four groups.
Exposure to diesel exhaust particulate matter in conjunction with consumption of a HF diet results in increased furin protein expression in the bronchioles of C57BL/6 mice. Representative images of furin expression in the bronchioles of C57Bl/6 mice on a control (LF; A-C) or high-fat (HF; G-I) diet exposed to saline (CON) or on a LF (D-F) or HF diet (J-L) exposed to diesel exhaust particles (DEP – 35 µg PM) twice a week for a total of 30 days. Red fluorescence indicates furin expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung furin mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of furin mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification; scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to LF + CON, †p < 0.05 compared to LF + DEP, ‡p < 0.05 compared to HF + CON by two-way ANOVA
Probiotic treatment mitigated the DEP- and HF diet-mediated increase in furin protein expression in the bronchioles of C57BL/6 mice
To determine whether probiotic treatment mitigates DEP- ± HF diet-mediated alterations in furin expression, we measured furin expression by immunofluorescent histology (Fig. 10A-M) and mRNA transcript expression by RT-qPCR (Fig. 10N). We observed a significant decrease in furin protein expression in the groups treated with probiotics [HF + CON + PRO (p < 0.0001), HF + DEP + PRO (p < 0.0001)] compared with the HF + DEP group. Furin protein expression was significantly lower in the HF + CON + PRO group than in the HF + CON (0.0360) and HF + DEP + PRO (p = 0.0077) groups. Exposure F = 46.02, probiotics F = 60.13, exposure x probiotics F = 12.00.
Furin mRNA expression was decreased in the HF + CON + PRO group compared with the HF + CON (p = 0.0399) and HF + DEP + PRO (p = 0.0022) groups. There was no significant difference observed in furin mRNA expression among the HF + CON, HF + DEP, and HF + DEP + PRO groups. Exposure F = 0.04313, probiotics F = 3.122, exposure x probiotics F = 7.989.
Probiotic treatment mitigates DEP- and HF diet-mediated increase of furin protein expression in the bronchioles of C57BL/6 mice. Representative images of furin expression in the bronchioles of C57Bl/6 mice on a high-fat (HF) diet exposed to either (A-C) saline (CON), (D-F) DEP – 35 µg PM, (G-I) saline and probiotics (PRO) − 0.3 g/day (~ 7.5 × 107 cfu/day), or (J-L) DEP and probiotics. Red fluorescence indicates furin expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung furin mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of furin mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification, scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to HF + CON, †p < 0.05 compared to HF + DEP, ‡p < 0.05 compared to HF + CON + PRO by two-way ANOVA














Add Comment