Materials
Zn(NO3)2·6H2O, sodium chondroitin sulfate, and 2,4-dimethylimidazolewere sourced from Shanghai Aladdin Biochemical Technology Co., Ltd. TP5 and MTX were provided by Meilun Biotech Co., Ltd. Thiazolyl blue tetrazolium bromide (MTT) was purchased from Aladdin Chemicals. The CT26 and RKO cells were provided by American Type Culture Collection (ATCC). DMEM/1640 culture medium was obtained from Gibco. FBS was purchased from BI. The Annexin V-FITC/PI and the TUNEL cell apoptosis detection and the cell apoptosis detection kits were provided by Yeasen Biotechnology Shanghai Co., Ltd. Enzyme-linked immunosorbent assay (ELISA) kits for mouse IL-2, mouse IL-6, mouse IL-10, mouse IFN-gamma (IFN-γ) and TNF-α were provided by ABclonal. The mouse granzyme B ELISA kit was purchased from Solarbio. Rabbit mAbs from Biolegend against the following were used: cGAS (79978), STING (13647), phospho-STING (AF7416), IRF-3 (4302 S), and phospho-IRF-3 (4302 S). Anti-GLUT1 rabbit mAb (bsm-52240R) was provided by Bioss Biotech Beijing Co. Ltd. (China). AMPK (ab72845), phospho-AMPK rabbit mAb (AP1002), β-actin rabbit mAb (AC050), and CD8A rabbit polyAb (A11856) were purchased from ABclonal. Anti-DFNA5/GSDME (ab215191) was provided by Abcam. PD-L1 rabbit mAb (M033179) was obtained from ABMART. Anti-caspase-3 (PTM-5752) and anti-cleaved-caspase-3 (PTM-77246) rabbit mAbs were purchased from Biolab. Ki67 rabbit polyAb (28074-1-AP) was provided by Proteintech. Anti-CD8 (GB114196) and Anti-CD4 (GB15064) antibodies were from Servicebio. The erythrocyte lysis buffer was provided by Beijing Solarbio. Beyotime provided the ATP detection kit.
Nanoparticle characterization
The Malvern Zetasizer Nano Analyzer was employed to detect the zeta potential and size distribution of different nanoparticles. Transmission electron microscopy (TEM) images were captured with an HT7800 TEM instrument (Hitachi, Japan). An ultraviolet-visible (UV-vis) spectrophotometer (UV-2700) was utilized to obtain the UV-vis spectra. A laser pointer (Senwei, China) was used to record the typical tyndall effect of nanoparticles. To conduct elemental analysis via X-ray photoelectron spectroscopy (XPS), a K-Alpha instrument obtained from Thermo Fisher Scientific was used, while data analysis was conducted through Avantage software from Thermo Fisher Scientific. The chemical structure of each samples was confirmed based on X-ray diffraction (XRD) by an X’Pert Pro MPD instrument and Fourier-transform infrared spectroscopy (FTIR) using a Thermo Fisher Scientific Nicolet iS5 instrument, both of which were integrated into the scientific compass platform (www.shiyanjia.com) for subsequent analysis. A microplate reader (Synergy H1, BioTek, Vermont, USA) was used to determine the ATP levels and cell viability. Fluorescence images, including images generated by EdU fluorescence labeling for cell proliferation, were acquired with an inverted fluorescence microscope produced by Olympus (Tokyo, Japan). The in-vivo biodistribution of CS/NPs was investigated with a small-animal imaging system (PerkinElmer, Massachusetts, USA). An automatic biochemical analyzer (Chemray 240, Rayto, China) was used to perform blood biochemical analysis. The drug release data and ELISA measurements were obtained using a Tecan-Spark multifunctional microplate reader (Tecan, Switzerland). Cellular uptake, flow cytometry, and immunological analyses were completed with a BD FACSCelesta Analytical Flow Cytometer (BD/Becton Dickinson, New York New Jersey, USA).
Synthesis of ZIF-8@MTX/TP5
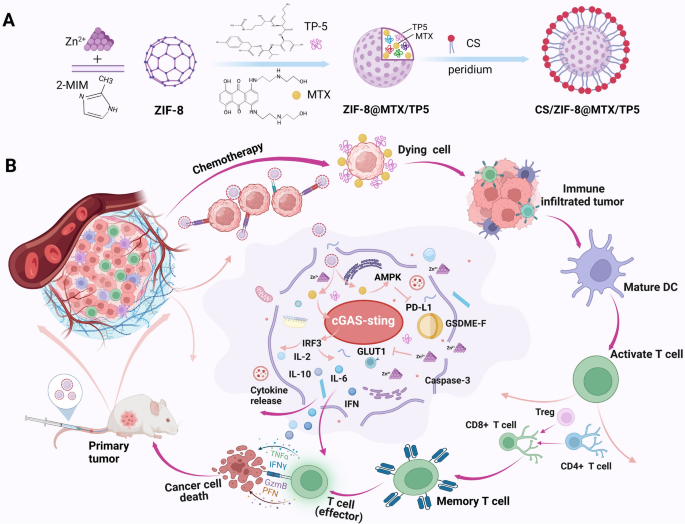
The synthesis of ZIF-8@MTX/TP5 NPs was accomplished following a one-pot method. First, to create solution A, both 40 mg MTX and 40 mg TP5 were dissolved in double-distilled water (ddH2O, 4 mL). In parallel, 100 mg zinc nitrate Zn(NO3)2·6H2O was dissolved in ddH2O (0.4 mL) to obtain solution B, while 1 g 2-methylimidazole (2-MIM) was dissolved in ddH2O (4 mL) to create solution C. Solution B was stirred at 600 rpm for 5 min, and solution A was then added dropwise while stirring for a further 10 min. The mixture was then added dropwise to solution C under continuous stirring at 800 rpm for 15 min. Subsequently, the product was obtained using centrifugation at 13,000 rpm for 30 min and rinsed three times in ddH2O.
Synthesis of CS/ZIF-8@MTX/TP5
To synthesize CS/ZIF-8@MTX/TP5, of ZIF-8@MTX/TP5 nanoparticles (100 mg) were suspended in of 3% (w/v) chondroitin sulfate (CS) solution (30 mL). The suspension was subjected to probe sonication for 30 min, after which magnetic stirring proceeded at 1000 rpm for 24 h. After centrifugation, the product was washed three times with ddH2O to obtain CS/NPs, as illustrated in Scheme 1A.
MTX and TP5 loading efficiency
The formula used to determine the loading efficiency of MTX and TP5 was as follows: Encapsulation efficiency (%) = WE/WT ×100, where WE is the quantity of MTX and TP5 encapsulated in CS/ZIF-8@MTX/TP5. WT represents the entirety of MTX and TP5 added. The amounts of MTX and TP5 were determined through UV-vis spectroscopy absorption based on the standard curves.
Cell culture conditions
The mouse-derived CRC cell line CT26 and the human-derived CRC cell line RKO was conducted in RPMI 1640 medium (Gibco) by adding 1% penicillin-streptomycin and 10% FBS. Cell culture proceeded in an incubator maintained at 37 °C in a 5% CO2 atmosphere.
Cellular uptake assay in vitro
To investigate the absorption of CS/NPs, RKO and CT26 cells were added to six-well plates (density: 2 × 104 cells/well). The culture medium was taken out, and the wells were provided with fresh medium containing 10 µM NPs. Cell incubation was then conducted for 0, 1, 2, 4, 6, and 8 h. Following incubation, cells were rinsed thrice in PBS for the removal of drug residue. The absorption was assessed via flow cytometry and inverted fluorescence microscopy to examine digested and fixed cells.
Cell viability assay
To assess the cytotoxicity of CS/NPs, an MTT assay was performed. RKO and CT26 cells were added to 96-well plates (density: 4 × 103 cells/well) and allowed to incubate overnight. After incubation, cells underwent 24 h of treatment with normal saline (Ctrl) and varying concentrations of drugs singly or in combination (TP5, MTX, MTX/TP5 (M + T), NPs, and CS/NPs). Supernatant collection and cell rinsing with PBS were conducted prior to the MTT assay using a microplate reader. In addition, the anti-proliferative ability of different treatments were further assessed using colony formation assays. Twenty-four-well plates were used for the assay, with RKO and CT26 cells seeded (density: 1 × 103 cells/well) and incubated for 3 days. After incubation, cells were subjected to treatments with Ctrl, TP5, MTX, (M + T), NPs, or CS/NPs. Following drug withdrawal, the cells were cultured in an incubator at 37 °C under a 5% CO2 atmosphere for 7 days. The resulting cell colonies were gently rinsed three times, fixed for 30 min in 4% paraformaldehyde (PFA), and stained with crystal violet. The colony numbers were captured with a digital camera, and ImageJ software was employed to perform counting. The colony formation rate (%) was calculated based on comparing the area of crystal violet staining in different groups.
EdU labeling assay
EdU labeling was explored using the EdU Cell Proliferation Assay Kit. In brief, CRC cells were added to 96-well plates (density: 5 × 103 cells/well) and subjected to treatment with various concentrations of Ctrl, TP5, MTX, M + T, NPs, and CS/NPs based on the drug uptake times. Following the removal of the supernatant and incubation for a further 24 h, photographs of the cells were captured with an inverted fluorescence microscope and counted with ImageJ software.
Cell apoptosis assay
The Annexin V-FITC/PI Cell Apoptosis Assay Kit was utilized to determine the rate of tumor cell apoptosis. Six-well plates were used to incubate cells seeded (density: 5 × 104 cells/well) for 24 h. Subsequently, cells were subjected to treatment with Ctrl, TP5, MTX, M + T, NPs, or CS/NPs and incubated for a further 24 h. Cells were then stained with both annexin V-FITC and PI (30 min) following the manufacturer’s protocol. Finally, flow cytometry was performed for the assessment of apoptosis in accordance with the manufacturer’s instructions.
Western blotting
After drug treatment, cells were taken via scraping, rinsed using cold PBS, and subjected to sonication in radioimmunoprecipitation assay (RIPA) buffer containing SDS (0.1%), sodium deoxycholate (1%), and Triton X-100 (1%), with added phosphatase inhibitors and protease inhibitors. SDS-PAGE was performed on the lysates, which were then transferred onto PVDF membranes. Next, the membranes were blocked at room temperature for 2 h using skimmed milk. Incubation was carried out at 4 °C with specific primary antibodies followed by 2 h incubation with secondary antibodies. Protein expression was visualized using Immobilon Western HRP Substrate and the ChemiScope 0050 Touch Integrated Chemiluminescence Imaging System.
ATP detection assay
ATP detection kits were employed to determine the level of ATP. In this assay, cells were seeded in six-well plates (density: 96 × 104 cells/well). After drug intervention, the cell culture supernatant was collected and processed. Then, 20 µL supernatant was mixed with a portion of the assay’s working solution (100 µL). A multifunctional enzyme marker was used to obtain the ATP fluorescence intensity.
Co-culture experiments
We validated the in vitro liposomal cell immunological results through co-culture experiments. Cell culture was performed in six-well plates (density: 5 × 104 cells/well). Following drug treatment, the cells underwent incubation for 24 h. Additionally, splenic cells were obtained from female BALB/c mice and seeded in the six-well plates. Co-incubation of tumor cells and splenic cells was conducted in an incubator at 37 °C for 48 h. The supernatant containing the cells was harvested and underwent centrifugation at 4000 rpm for 3 min. Following centrifugation, the supernatant was disposed of and the cells were obtained. The cells in the six-well plates were treated with trypsin and the digestion was terminated with a complete culture medium. Following centrifugation at 4000 rpm for 3 min, the supernatant was discarded and the cells were collected. The cells underwent staining with anti-CD11c-PE, anti-CD80-FITC, anti-CD86-APC, anti-CD3-FITC, anti-CD4-PE, and anti-CD8-APC antibodies, and flow cytometry was employed to determine the percentage of mature DCs and T-cell activation.
In vivo imaging and biodistribution analysis
To assess drug accumulation in the tumor area, the drugs were intravenously administered to Luc-CT26 subcutaneous tumor-bearing mice when the tumor volume reached 150–200 mm3. First, free single-drug MTX (1 mg kg−1), non-targeted ZIF-8@MTX/TP5 (2 mg kg−1), and CS/ZIF-8@MTX/TP5 (2 mg kg−1) were injected into the tail vein. Mice underwent anesthesia with 3% isoflurane, and images of mice were captured with an in-vivo imaging system (IVIS Lumina XR III, PerkinElmer, Massaachusetts, USA) at 1, 2, 4, 6, 8, and 24 h post-injection. Mice were euthanized for biodistribution analysis following the 24-h imaging, and tumors and major organs were dissected and used to obtain ex-vivo fluorescence images.
In vivo antitumor study
Yaokang Biotechnology Co., Ltd. (Chengdu, China) supplied six-week-old female BALB/c mice that weighed approximately 18–20 g. All the animals studies were conducted in accordance with the “Guidelines for Animal Experiments” and were approved by the Sichuan University Animal Ethics and Use Committee. First, a unilateral subcutaneous tumor model was created through the injection of mice with Luc-CT26 colorectal cancer cells (5.0 × 105 cells suspended in serum-free 1640 medium) in the right axillary subcutaneous region. When the volume of the tumors had grown to 100 mm3, mice were selected at random and split into six groups (n = 5 per group) to explore the antitumor effects of the multifunctional NPs. All drug treatments, namely, Ctrl (normal saline), TP5, MTX, (M + T), NPs, and CS/NPs (2 mg/kg), were administered to mice by tail vein injection every other day. The tumor volume and body weight of mice were recorded every 2 days. A vernier caliper was used to obtain the tumor size, and the volume of tumors (mm3) was determined by the following formula: \(\:\text{T}\text{u}\text{m}\text{o}\text{r}\:\text{V}\text{o}\text{l}\text{u}\text{m}\text{e}\:\left({\text{m}\text{m}}^{3}\right)=\frac{\text{L}\text{e}\text{n}\text{g}\text{t}\text{h}\times\:{\text{W}\text{i}\text{d}\text{t}\text{h}}^{2}}{2}\). On day 15 of therapeutic intervention, all mice were euthanized and tumor tissue were removed and weighed to assess the antitumor effect of drug treatments. The tumor inhibition rate was calculated according to the following equation:
$$\:\text{I}\text{n}\text{h}\text{i}\text{b}\text{i}\text{t}\text{i}\text{o}\text{n}\:\text{R}\text{a}\text{t}\text{e}\:\left(\%\right)\frac{\text{T}\text{u}\text{m}\text{o}\text{r}\:\text{w}\text{e}\text{i}\text{g}\text{h}\text{t}\:\text{i}\text{n}\:\text{c}\text{o}\text{n}\text{t}\text{r}\text{o}\text{l}\:\text{g}\text{r}\text{o}\text{u}\text{p}-\text{T}\text{u}\text{m}\text{o}\text{r}\:\text{w}\text{e}\text{i}\text{g}\text{h}\text{t}\:\text{i}\text{n}\:\text{t}\text{r}\text{e}\text{a}\text{t}\text{m}\text{e}\text{n}\text{t}\:\text{g}\text{r}\text{o}\text{u}\text{p}}{\text{T}\text{u}\text{m}\text{o}\text{r}\:\text{w}\text{e}\text{i}\text{g}\text{h}\text{t}\:\text{i}\text{n}\:\text{c}\text{o}\text{n}\text{t}\text{r}\text{o}\text{l}\:\text{g}\text{r}\text{o}\text{u}\text{p}}\times\:100\%.$$
The tumors and spleen, heart, kidneys, liver, lung tissue were further harvested for subsequent experiments.
Anti-tumor immune activation experiment
Tumor tissue and spleens were collected from the treated mice at the 15th day, and grinding and processing were used to prepare single-cell suspensions. Cell staining was conducted using anti-CD11c-PE, anti-CD3-FITC, anti-CD4-APC, anti-CD80-FITC, anti-CD4-PE, anti-CD86-APC, anti-CD8-APC anti-CD25-FITC, and anti-Foxp3-PE antibodies, after which flow cytometry was performed.
The activation of immune response
In this study, a bilateral subcutaneous tumor model was established using BALB/c mice through the intratumoral injection of CT26-Luc cells (5 × 105 cells) to induce the growth of proximal primary tumors in the left axillary subcutaneous region of female BALB/c mice coupled with the subcutaneous injection of CT26-Luc cells (5 × 105 cells) into the right axillary subcutaneous region of the same mouse to induce the growth of distal tumors. When the volume of tumors had grown to approximately 6 mm, mice were selected at random and placed into one of six groups: Ctrl (control), TP5, MTX, (M + T), NPs, and CS/NPs (n = 5 per group). Peritumoral drug administration was conducted on alternating days, with the tumor volume and body weight of each mouse recorded every 2 days. On day 14, bilateral tumor tissues and spleens were excised from treated mice and processed to generate single-cell suspensions. Tumor cells underwent staining with anti-CD11c-FITC, anti-CD80-APC, anti-CD86-PE, anti-CD3-FITC, anti-CD4-PE, anti-CD8-APC, anti-CD4-APC, anti-CD25-FITC, and anti-Foxp3-PE antibodies, after which flow cytometry was performed to obtain the proportions and numbers of CD4+ and CD8+ T cells in the proximal left and distal right tumors and spleens.
Histological staining
The harvested mouse tumors and major organs were fixed using 4% PFA and then embedded in paraffin. Tissue sections with a thickness of 4 μm were taken from paraffin-embedded blocks and histological examination was performed based on hematoxylin and eosin (H&E) staining. In addition, immunohistochemical staining (IHC) was conducted for KI67, PD-L1, cGAS, and P-STING to evaluate protein expression levels in tumor tissues.
Immunofluorescence
First, cells were placed onto glass coverslips in 24-well plates (density: 5 × 103 cells/well) and left to adhere for 24 h. After 24 h, the cells were fixed in 4% PFA, rinsed using PBS, and permeabilized with 0.4% Triton X-100. The cells were then blocked with 5% FBS and incubated with primary antibodies specific for CD4 and CD8. Following incubation with primary antibodies, the cells underwent treatment with CY3-labeled goat anti-rabbit IgG secondary antibodies. Cellular images were captured using a scanner (Pannoramic MIDI, 3DHISTECH, Hungary).
ELISA assays
ELISA assays were conducted using specific ELISA kits. Mouse sera were collected, and the concentrations of cytokines including TNF-α, IFN-γ, IL-1, IL-2, IL-6, IL-10, and granzyme B were determined using the respective ELISA kits. Concentration measurements were analyzed using the Tecan-Spark Multimode Microplate Reader.
Serum biochemical analysis
Biochemical assay kits were employed to analyze the mouse sera. The concentration of aspartate aminotransferase (AST), alanine aminotransferase (ALT), creatinine (CRE), and urea, which serve as markers of kidney and liver function, were determined with an automated biochemical analyzer.
Statistical analysis
We used GraphPad Prism 8.0 software for statistical analyses, which were performed using one-way analysis of variance (ANOVA) or t-tests. The related data were shown as the Mean ± Standard Deviation (SD) of at least 3 separate assays. *mean p < 0.05; ** mean p < 0.01; *** mean p < 0.001; NS mean not significant.



Add Comment