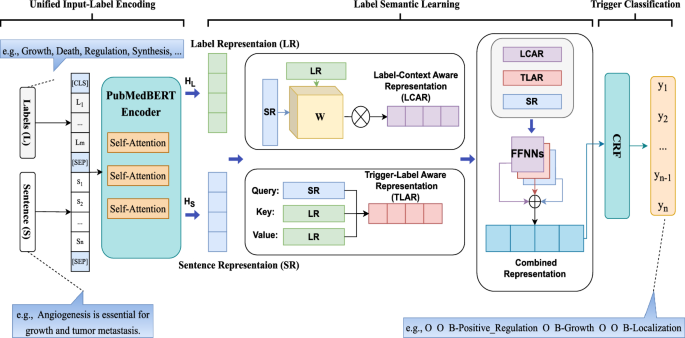
Figure 3 shows the overall architecture of the proposed Biomedical Label-based Synergistic representation Learning (BioLSL) model for biomedical event trigger detection. The proposed BioLSL model takes in a sentence S in the form of \(\{s_1, s_2,…, s_n\}\), where \(s_i\) is the i-th token in the sentence and n denotes the total number of tokens, and \(T_{BED} = \{t_1, t_2,…, t_k\}\) is a set of pre-defined event type labels, where k denotes the total number of event types. The model outputs a predicted label sequence L in the form of \(\{l_1, l_2,…, l_n\}\), where \(l_i\) denotes the type label for \(s_i\), and \(l_i \in T_{BED}\). Moreover, we use the BIO tagging scheme [47] in our model to mark the trigger words consisting of multiple tokens. The BIO tagging scheme is a tagging format for representing labels in a sequence, which uses the following three tags: “B” (beginning of the event trigger), “I” (inside of the event trigger) and “O” (outside the event trigger). As such, it can maintain the boundaries between adjacent event triggers. For example, given the sentence “Angiogenesis is essential for growth and tumor metastasis.”, the model produces the output {O, O, B-Positive_Regulation, O, B-Growth, O, O, B-Localization} according to the BIO tagging scheme.
The proposed BioLSL model consists of three modules, namely Domain-specific Joint Encoding, Label-based Synergistic Representation Learning and Trigger Classification.
Domain-specific joint encoding
This module takes in the input sentences and a list of pre-defined type labels for encoding. In particular, the biomedical pre-trained language model PubMedBERT [21] is used to encode each word in the input sentence, which is represented as:
$$\begin{aligned} X_{L,S} = \langle [CLS],L,[SEP_1],S,[SEP_2] \rangle \end{aligned}$$
(1)
where L is the type label words, S is the input sentence, and [CLS] and [SEP] are special tokens in PubMedBERT. Note that L is a fixed list of text names describing the pre-defined event types’ concepts. For example, the MLEE [3] biomedical corpus contains 19 pre-defined event types such as Growth, Death, Regulation, etc. and L is created as a random sequence of type labels, e.g., {Growth, Death, Regulation, Synthesis,… } . For the few type labels whose names contain multiple words, we employ additional special tokens to represent them.
Inspired by [20], the Domain-specific Joint Encoding module uses PubMedBERT’s multi-head self-attention mechanism [48] to capture the direct interactions between the type labels L and the input sentence S. Then, the representation for the type labels and the sequential representation for input sentence tokens are generated through the multiple Transformer layers. The attention heads in the Transformer layers can be expressed as follows:
$$\begin{aligned} ATTENTION(Q,K,V)=softmax\left( \frac{QK^T}{\sqrt{d_k}}\right) V \end{aligned}$$
(2)
where \(d_k\) denotes the encoding dimension, and Q, K, V represent the query, key, and value matrices, respectively.
Then, PubMedBERT is used to generate the representations for the type labels and the input sentence:
$$\begin{aligned} (H_L,H_S) = PubMedBERT(X_{L,S}) \end{aligned}$$
(3)
where \(H_L\) is the encoded token sequence of type label words, and \(H_S\) is the encoded token sequence of the input sentence, called Label Representation (LR) and Sentence Representation (SR), respectively. These two representations are derived by splitting the output representation from PubMedBERT.
Label-based synergistic representation learning
In biomedical event trigger detection, predicting event triggers is highly context-dependent. For example, as pointed out by Pyysalo et al. [3], a term is considered indicative of a Growth event only when it can distinctively make reference to the “upper-level gene ontology”. To capture such contextual information, it is essential to capture the meaningful contextual input sentence tokens and match them with their corresponding type label tokens. Therefore, we first obtain the Label-Context Aware Representation (LCAR), C, that captures the intricate interaction of the contextual input sentence tokens with the event types.
The label-context aware representation \(C_{i}\) of input sentence token \(S_i\) is computed as follows:
$$\begin{aligned} \begin{aligned} C_{i} = \frac{1}{N} \sum _{j=1}^N {\tilde{a}}_{ij} \cdot h_L^{(j)} \\ {\tilde{a}}_{ij} = \theta (h_S^{(i)},h_L^{(j)}) \end{aligned} \end{aligned}$$
(4)
where N is the total number of type labels, and \(\theta (.)\) denotes the attention function, which is computed using the input sentence representation for query, and the type label representation for both key and value.
After that, to determine whether an input sentence token \(h_S^{(i)}\) is the candidate trigger of a certain event type, we need to calculate the semantic proximity and capture the underlying semantic relationship between the latent trigger tokens in input sentence and the target type label tokens. Therefore, we compute an interaction matrix, W, which links the encoded token sequence of type label words (i.e., \(H_L \in {\mathbb {R}}^{m \times d_k}\)), with an encoded token sequence of the input sentence (i.e., \(H_S \in {\mathbb {R}}^{n \times d_k}\)). Here, m represents the total number of type label words, n refers to the number of tokens in the input sentence, and \(d_k\) indicates the encoding dimension. Then, it generates the Label-Trigger Aware Representation (LTAR), \(A_{i,j}\), for each pair of a type label and an input sentence token \(<S_i,L_j>\) as follows:
$$\begin{aligned} A_{i,j} = \sigma ({h_S^{(i)}}^\top W h_L^{(j)} + b) \end{aligned}$$
(5)
where \(\sigma\) denotes the sigmoid nonlinearity function and b is a bias term. The interaction matrix W is learnable and continues to update during training. Thus, the matrix can capture the underlying semantic relationships between the type labels and their corresponding trigger words.
Besides the original input sentence representation (i.e., \(H_S\)), our proposed model also contains the semantic information from the label-context aware representation and label-trigger aware representation. These two representations learn latent information from label-trigger and label-context relationships to help the detection of biomedical event triggers. We map these three representations (i.e., \(H_S\), C and A) to a Conditional Random Field (CRF) [22] for decoding by using three separate single layer feed-forward neural networks (FFNNs) with cross-type activation functions:
$$\begin{aligned} \begin{aligned} X’&= FFNN_{1}(H_S) \\ {\hat{X}}&= FFNN_{2}(C)\\ {\tilde{X}}&= FFNN_{3}(A) \end{aligned} \end{aligned}$$
(6)
where \(X’\), \({\hat{X}}\), and \({\tilde{X}}\) represent the corresponding mapped representations, with each being a matrix in \({\mathbb {R}}^{n \times k}\), where k refers to the number of type labels based on the BIO tagging scheme. Most previous works [49] concatenate different representations for joint decoding. However, it may potentially lead to overly sparse feature representations and cause the problem on gradient vanishing. As concatenation increases the dimensionality of the feature space, it will render the features more sparsely in the high-dimensional space [50]. The sparsity poses a significant challenge to the model’s learning capability, as the model will struggle to learn patterns from the sparse features and generalize them accordingly. This is particularly problematic in biomedical event trigger detection, where the number of training samples is relatively small compared to the vast feature space [51]. Therefore, we use different FFNNs to map each representation individually.
We combine the separate representations by applying a weight parameter to balance each representation’s contribution. Drawing inspiration from the work on residual learning [52], we aim to ensure the preservation of valuable information and avoid potential model degradation. In particular, as discussed in [53, 54], the sentence representation output from BERT carries important contextual information. Therefore, we balance and combine the different representations, denoted as \(x = \{x_1,x_2,…,x_n\}\), as follows:
$$\begin{aligned} x_i = x’_i + \alpha {\hat{x}}_i + (1-\alpha ) {\tilde{x}}_i \end{aligned}$$
(7)
where \(x’_i \in X’\), \({\hat{x}}_i \in {\hat{X}}\), and \({\tilde{x}}_i \in {\tilde{X}}\). And \(\alpha \in (0,1)\) is a hyperparameter to be determined empirically.
Trigger classification
This module uses CRF for identifying event trigger candidates by decoding the combined representation and predicting the event trigger. Since our proposed model makes use of the BIO tagging scheme for modeling the type labels, it is important to consider label sequence. For example, the label “I” should not directly follow the label “O”. Activation functions such as Softmax are unable to take label dependencies and label sequence into consideration for prediction.
Given the combined representation x obtained from the Label-based Synergistic Representation Learning module, CRF computes the probability of a ground truth type label sequence \(y= \{y_1,y_2,…,y_n\}\) as follows:
$$\begin{aligned} P(y\mid x)= & {} \frac{exp \big ( score(x,y) \big )}{\sum _{y’ \in Y} exp \big ( score(x,y’) \big )} \end{aligned}$$
(8)
$$\begin{aligned} score(x,y)= & {} \sum _{i=0}^{n-1} T_{y_i, y_{i+1}} + \sum _{i=0}^{n} F_{x,y_i} \end{aligned}$$
(9)
where n is the length of x, Y is a set of all possible type label sequences, and \(y’\) is the predicted type label sequence. T is the transition matrix, with \(T_{y_i,y_{i+1}}\) being the transition parameter from label \(y_i\) at position i to label \(y_{i+1}\) at position \(i+1\). \(F_x\) is the emission matrix of the representation x, with \(F_{x,y_i}\) being the score of label \(y_i\) at position i with respect to x.
After that, we use the Negative Log-Likelihood loss function [55] to measure the distance between the predicted type label sequence and the true type label sequence:
$$\begin{aligned} {\mathcal {L}}(x, y) = -\log P(y|x) \end{aligned}$$
(10)
where y is the ground true type label sequence and x is the combined representation. Finally, our proposed BioLSL model minimizes the loss function \({\mathcal {L}}(x, y)\) during training by optimizing the parameters of the proposed model.
For prediction, we apply the argmax function to the probability distribution \(P(Y\mid X_{L,S})\) to obtain the predicted type label sequence \(y^*\) as follows:
$$\begin{aligned} y^* = \mathop {argmax}P(Y \mid X_{L,S}) \end{aligned}$$
(11)





Add Comment