Identification and resistance phenotypes
Clinical E. coli strains (ECO20 and ECO21) were isolated from hospitalized patients at a hospital in Jining, Shandong. Notably, both strains exhibited the presence of both blaNDM−7 and mcr-1 genes. ECO20 was recovered from the urine sample of Patient 1 in the intensive care unit on February 17, 2021, while ECO21 was obtained from Patient 2’s sputum in the neurology department on February 19, 2021 (Figure S1). Despite their non-concurrent presence in the same ward, clinical records reveal a compelling temporal and spatial correlation. Patient 2, carrying ECO21, had a prior admission to the ICU ward from February 09 to February 18, 2021 (Figure S1). During an overlapping interval (February 15–18), both Patient 1 and Patient 2 coexisted within the ward, suggesting relevant temporal and spatial proximity influencing transmission dynamics.
Antimicrobial susceptibility testing revealed that both E. coli isolates conferred resistance to almost antimicrobials tested, including ampicillin, ampicillin/sulbactam, piperacillin/tazobactam, cefazolin, cefuroxime, ceftriaxone, cefepime, cefoperazone/sulbactam, gentamicin, trimethoprim/sulphamethoxazole, imipenem, meropenem, and colistin, but no resistance to tigecycline, nitrofurantion, and amikacin (Table 1). Thus, these strains exhibited multidrug-resistant profiles.
Genomic characteristics
To further investigate the clonal relationship and genomic features, we sequenced and assembled their genomes. Notably, the ECO20 genome shared 99.99% Average Nucleotide Identity (ANI) with ECO21 (Figure S2), indicating the clonal transmission of these two isolates. The two strains belonged to ST4456, fimH20 and H4:O83, and harboring some common virulence genes such as csgA, fdeC, hlyF, ibeA, iss. They exhibited identical plasmid profiles: a 33 Kb pMCR plasmid, a 46 Kb pNDM plasmid, and a 175 Kb pVir plasmid (Table 2). The pMCR plasmid belonging to the IncX4 type carried the colistin resistance gene mcr-1, while the pNDM plasmid belonging to the IncX3 type carried the carbapenem resistance gene blaNDM−7. The pVir plasmid (IncFIB/IncFII type) harbored aerobactin and salmochelin-associated virulence genes, alongside additional resistance genes, including AAC(3)-IId, dfrA17, TEM-1, sul2, APH(3’’)-Ib, APH(6)-Id, and tet(A). Notably, all plasmids possessed conjugal transfer elements, albeit the oriT sequences of pNDM and pMCR were not identified by the oriTfinder web tool.
Transferability of resistance and virulence plasmids
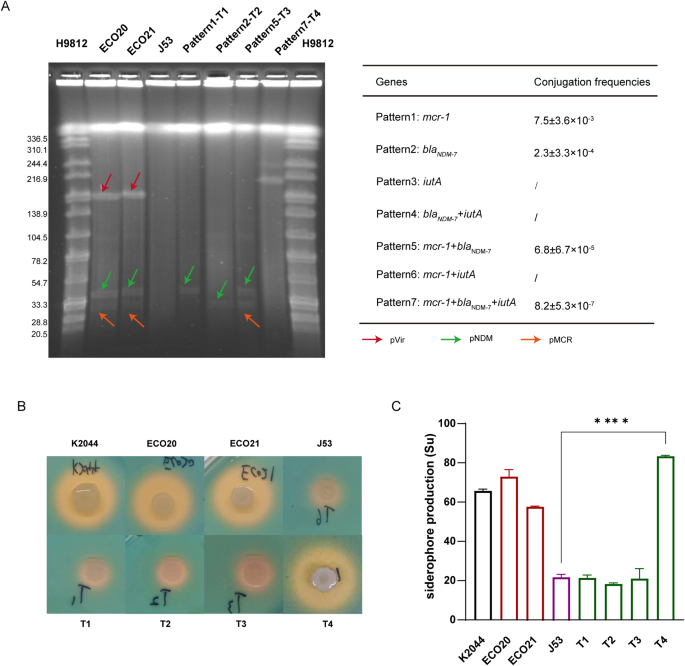
To investigate the transmissibility of mcr-1, blaNDM−7, and iutA, we subjected ECO20 to conjugation experiments with E. coli J53. As shown in Fig. 1A, there should be seven plasmid conjugation patterns. However, after many conjugation experiments, we only observed four different transfer patterns (Fig. 1A). Importantly, the co-transfer of mcr-1, blaNDM−7, and iutA led to the formation of a fusion plasmid (Fig. 1A), suggesting that diverse recombination events would occur during the plasmid conjugation process. High conjugation frequencies of mcr-1 and blaNDM−7 indicate broad spread and effective transfer of pMCR and pNDM plasmids (Fig. 1A). The transfer of resistance genes mcr-1 and blaNDM−7 conferred their corresponding resistance phenotypes against colistin and carbapenem to E. coli J53 (Table 1). Similarly, virulence genes with their corresponding virulence phenotypes characterized by high siderophore production were also successfully transferred to E. coli J53. The transconjugant T4 which acquired the pVir plasmid from ECO20 showed significantly higher siderophore production than J53 (Fig. 1B and C).
Prevalence of pMCR and pNDM plasmids
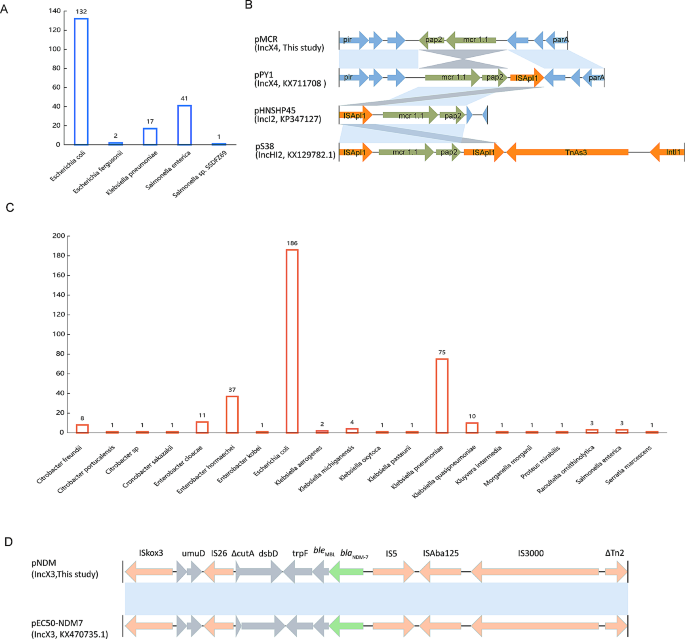
Blastn analysis revealed that the IncX4-type pMCR plasmid closely resembled numerous plasmids previously submitted to the NCBI database (Figure S3). These similar plasmids exhibited >99% coverages and >99% identities. Interestingly, it was not confined to E. coli but was also found in Klebsiella and Salmonella (Fig. 2A). Moreover, the single-ended Tn6330 variant (ISApl1–mcr1.1-pap2) is commonly present in mcr-1.1-harbouring plasmids, such as pHNSHP45(IncI2, KP347127) and pS38(IncHI2, KX129782.1), despite that ISApl1 was downstream of mcr1.1 and pap2 in pPY1(IncX4, KX711708) (Fig. 2B). In contrast, pMCR in this study had a different genetic environment for mcr1.1 with the pap2 located downstream and ISApl1 deleted (Fig. 2B).
The pNDM-like IncX3 plasmid was also identical to many plasmids from the NCBI database (Figure S4). These similar plasmids had 100 coverages and > 99% identities. They were frequently observed in gram-negative bacteria, exhibiting a broader host range compared to the pMCR-like IncX4. Apart from E. coli and Klebsiella, the pNDM-like IncX3 plasmid was also detected in Enterobacter, Citrobacter, and various other bacterial species (Fig. 2C). Nonetheless, E. coli was still the primary host bacterium carrying the blaNDM−7-positive IncX3 plasmid (Fig. 2C). Unlike the prevalence observed in pMCR-like IncX4, the incidence of pNDM-like IncX3 plasmids was lower in Salmonella (Fig. 2C). Furthermore, blaNDM−7 was associated with the complex transposon structure (IS26-IS3-ISAba125-IS5-blaNDM−7– bleMBL-trpF-dsbD-IS26) which is also commonly present in other blaNDM−7-positive plasmids and plasmids carrying NDM variants (Fig. 2D).
Characteristics of the IncFIBK/IncFII virulence plasmid
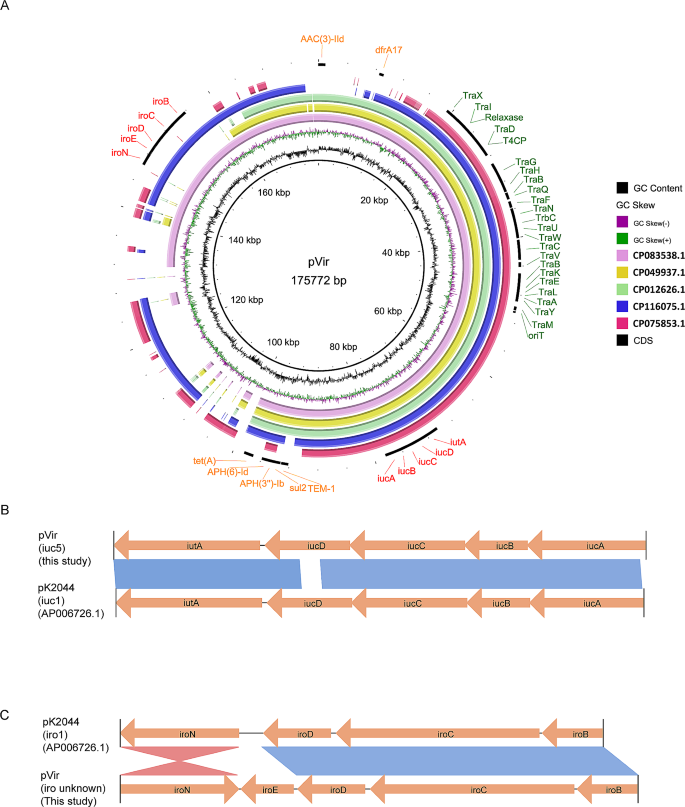
The 175,772 bp pVir plasmid contained virulence and resistance genes, along with a Type IV Secretion System (T4SS) (Fig. 3A). The aerobactin genes (iucABCD-iutA) and salmochelin genes (iroBCDEN) endowed it with high siderophore production (Fig. 1B and C). Notably, the schematic structure of iuc clusters (iuc1) was similar to that of virulence plasmid pK2044 in Klebsiella (iuc1) (Fig. 3B), while the iro clusters in pVir had iroE added and iroN inversion compared to pK2044 (Fig. 3C). The complete conjugal transfer element T4SS conferred the ability of self-transfer (Fig. 2A), and some resistance genes conferred resistance to aminoglycosides, cephalosporins, sulfonamides, tetracyclines, and diaminopyrimidines (Table 1). No identical plasmid was found in the NCBI nucleotide database. Some similar plasmids were selected to perform comparative genome analysis with pVir in this study. Most plasmids cannot possess all the above elements for self-transfer, resistance, and siderophore production, except for the unnamed plasmid (CP083538) (Fig. 3A).
Characteristics of siderophores-encoding pVir plasmids. (A) Comparative analysis of pVir plasmids and some similar plasmids from NCBI database. The reference plasmid is pVir (CP139892.1). (B) Comparisons of iuc and iro clusters carried by E. coli pVir plasmid and Klebsiella pneumoniae pK2044 plasmid





Add Comment