Comprehensive identification of ZoMYB gene family in ginger
A total of 231 ZoMYBs were identified in Z. officinale after removing redundant repetitive sequences. These genes, now designated ZoMYB#1 through ZoMYB#231, were chromosomally mapped and systematically renamed in accordance with their chromosomal positioning. The ZoMYB family is categorized into three distinct subfamilies: 156 R2R3-MYB proteins (2R-MYB sub family), 1 R1R2R3-MYB protein (3R MYB subfamily), and 74 MYB-related proteins (MYB related subfamily). Notably absent were representatives of the 4R-MYB class. Detailed attributes including coding sequence, protein sequence, sub-cellular localization, isoelectric point (PI), and molecular weights (MWs) are meticulously compiled in Supplementary Tables S1. ZoMYB#49 (Maker00069396) encodes the largest protein sequence with 1649 amino acid (aa), while ZoMYB#97 (Maker00078016) encodes the most compact protein at 73 aa, reflective of its singular MYB DBD domain. The molecular weights of the proteins span from 8243.9 Da (ZoMYB#97) to 182,772.28 Da (ZoMYB#49), with isoelectric points ranging from 4.38 (ZoMYB#97) to 9.96 (ZoMYB#215), respectively.
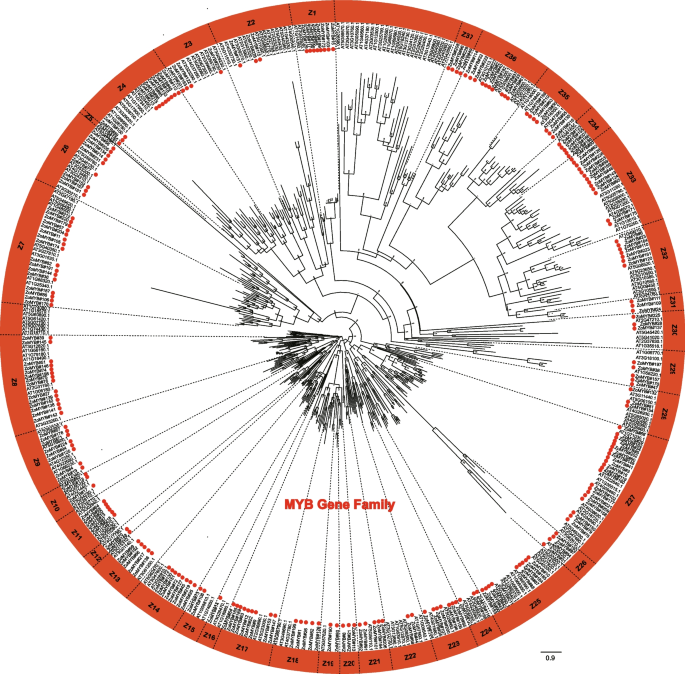
Phylogenetic insights into MYB genes in ginger and Arabidopsis
A phylogenetic reconstruction was undertaken using the amino acid sequences of the full complement of 231 ZoMYB and 197 AtMYB proteins to elucidate evolutionary relationships. Based on the phylogenetic tree, 231ZoMYB genes in ginger were divided into 37 groups (Z1-Z37) (Fig. 1, Supplemetary Document S1). Relying on the group classification of R2R3-MYB in Arabidopsis, the R2R3-MYBs were mainly belong to groups, designated Z2, Z3, Z6, Z7, Z8, Z9, Z10, Z11, Z12, Z13, Z14, Z15, Z17, Z18, Z20, Z21, Z22, Z23, Z24, Z25, Z27, Z28, Z32, Z33, and Z34. The MYB-related genes were mainly belong to Z1, Z26, Z29, Z30, Z31, Z35, Z36, and Z37. Notably, clades Z7, Z9, Z13, Z14, Z18, Z19, Z21, Z22, Z23, Z25, Z31, and Z32 have both MYB-related and R2R3-MYB genes. For example, clade Z6 included two MYB related type proteins, ZoMYB#56 and ZoMYB#77, together with other 7 R2R3 MYB proteins (ZoMYB#134, #152, #167, #181, #205, #231, and #214). Clade Z27, with 21 members, emerged as the most populous, while clades Z5 and Z16 were the least diverse, each containing a single membe.
Divergence in gene structure and motif composition within ZoMYBs
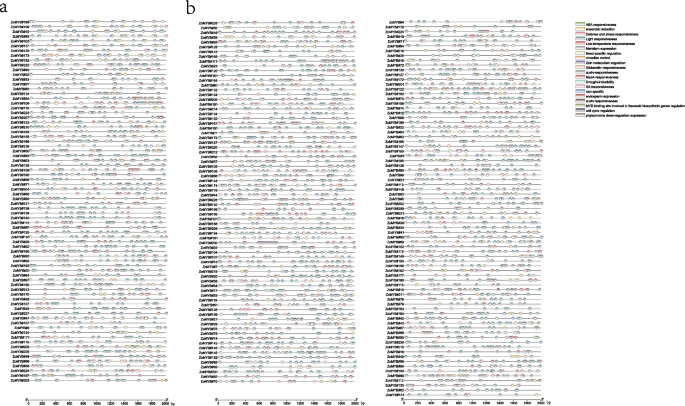
An intricate examination of the exon–intron architecture of ZoMYB transcription factors (TFs) in ginger was undertaken to elucidate the sequence diversity inherent to this gene family. The sequence structure of 1R-MYB, R1R2R3-MYB and R2R3-MYB in ginger were demonstrated separately. Illustrated in Fig. 2a, the 74 genes of the 1R-MYB category alongside a singular R1R2R3-MYB gene exhibited a range of exon counts, extending from a minimum of two to a maximum of 19. The intricacies of this exon–intron structural variance among the 74 1R-MYB genes, as well as the sole R1R2R3-MYB gene, are depicted in Fig. 2a. The exon tally for the 1R-MYB genes varied from one to 16, with the lone R1R2R3-MYB gene (ZoMYB#197) comprising a total of 19 exons. A notable proportion of the 1R-MYB genes predominantly possessed two (25.68%, 19/74) or three exons (29.73%, 22/74), while configurations of 10, 11, 15, and 16 exons were unique occurrences. As delineated in Fig. 4a, the ensemble of 156 R2R3-MYB genes was characterized by a disparity in exon numbers, which spanned from one to 11. A majority of the R2R3-MYBs in ginger conformed to the canonical splicing pattern of three exons (60.26%, 94 of 156 R2R3-MYB) interspersed by two introns (20.51%, 32/156). The CDS of most ZoMYB genes were interrupted by introns, with the exception of six ZoMYBs genes (ZoMYB#47, ZoMYB#122, ZoMYB#151, ZoMYB#163, ZoMYB#171, ZoMYB#201) which remained uninterrupted. In addition, when viewed in conjunction with the phylogenetic tree classification, members of the ZoMYB gene family that clustered within the same phylogenetic clade generally shared similar or identical exon counts, notwithstanding variations in exon positioning. For instance, the ZoMYB gene groups Z19 and Z20 both contained three exons, whereas the Z8 clade exhibited a range of two to six exons (Supplementary Table S1). In sum, the exon numbers of ZoMYB genes from ginger are quite divergent, however, the closer phylogenetical relationship is indicative of a greater homogeneity in sequence structure.
Distributions of gene structure and conserved motifs in 1R-ZoMYB and 3R-ZoMYB genes. a Exon/Intron structures of 1R-and 3R-ZoMYB genes. Green boxes represent untranslated regions (UTRs), yellow boxes denote exons, and black lines indicate introns. b Conserved motifs in 1R-and 3R-ZoMYB proteins, numbered 1–10 and depicted in various colors, with sequence details provided in Supplementary Figure S1. Scale at the bottom approximates protein length
To illuminate the conserved domains within ZoMYB proteins, motif analysis via MEME suite was performed, identifying twenty motifs across the 1R-MYB, R1R2R3-MYB and R2R3-MYB proteins of ginger (Figs. 3b, and 4b). As shown in Fig. 3b and Supplementary Figure S2, motif 2 and motif 6 were ubiquitous across the 1R-MYB proteins, whereas motif 1, 2, 3, and 7 were integral in encoding the MYB DNA-binding domain (DBD) within the R2R3-MYB proteins. In 1R-MYB types, different groups had different motifs, and motif 2 was common in all 1R-MYB proteins in Fig. 3b. The members from groups Z1 group were characterized by the presence of 9, 5, 7, 4, 8 and 2, while groups Z34 and Z35 harbored motifs 2, 6, and 1. Group Z31 was distinguished by motifs 5, 2, 6, and 1, and group Z8 by motifs 2, 6, and 3. This motif distribution underscores the lower sequence similarity among 1R-MYB proteins across different phylogenetic clades. Within the subset of 156 R2R3-MYB proteins, 85 were found to contain motifs 4, 3, 7, 1, and 2. ZoMYB#131 was exclusively composed of motifs 3 and 7. A unique motif signature, comprising motifs 6, 3, and 5, was identified in the R2R3-MYB proteins associated with the Z33 and Z32 clades. The 3R-MYB (ZoMYB#197) has three conserved motifs 2, 5 and 6, with motifs 2 repeated four times, and motifs 6 and 5 each being duplicated within this protein structure. Moreover, specific motif patterns unique to ZoMYB were discerned. For instance, ZoMYB#31 and ZoMYB#84 were solely characterized by motifs 2 and 5, respectively.
Distributions of gene structure and conserved motifs in R2R3-ZoMYB genes. a Exon/Intron structures of R2R3-ZoMYB genes. Green boxes, yellow boxes, and black lines indicate the 5′- and 3′-untranslated regions (UTR), exons, and introns, respectively. b Conserved motifs of R2R3-ZoMYB proteins. The motifs, numbers 1–10, denoted by a different color, is accompanied by sequence information in Supplementary Figure S3. The protein length can be estimated using the scale at the bottom
Analysis of cis-acting elements of ZoMYB promoters
A total of 57 cis-acting elements were identified in the promoter region (upstream 2000 base pair) of ZoMYBs, which could be classified into five categories: phytohormones (ABA responsiveness elements, Auxin responsiveness elements, Gibberellin responsiveness elements, MeJA responsiveness elements, SA responsiveness elements), environmental stress (Drought-inducibility elements, Defense and stress responsiveness elements, Low-temperature responsiveness elements, Anaerobic induction elements), photo-responsive elements (Light responsiveness elements, phytochrome down-regulation expression elements), growth and developmental elements (Cell cycle regulation elements, Circadian control elements, Root specific elements, Seed-specific regulation elements, Endosperm expression elements, Meristem expression elements) and secondary metabolic elements (MYB binding site involved in flavonoid biosynthetic genes regulation, Zein metabolism regulation), as delineated in Fig. 4, Supplementary Table S2, and Supplementary Table S3. The majority of ZoMYB genes exhibite the presence of a minimum of one phytohormone-responsive element within their promoter regions. A total of twelve phytohormone-responsive cis-acting elements were discerned, including ABRE, AuxRR-core, TGA-element, TGA-box, P-box, GARE-motif, TATC-box, TGACG-motif, CGTCA-motif, TCA-element, SARE, and O2-site. The cis-acting elements play a crucial role in regulating the responsiveness to ABA, auxin, gibberellin, methyl jasmonate, salicylic acid, and zein metabolism. Meanwhile, the ABRE responsiveness elements were the most common in the ZoMYB gene promoters. Furthermore, 34 cis-acting elements play a role in light responsiveness. Eight cis-regulatory elements associated with response to abiotic stresses were identified, including the low-temperature responsive element (LTR), anaerobic induction elements (ARE), drought-inducibility element (MBS), defense and stress responsive element (TC-rich repeats), and circadian control element (circadian).The Light responsiveness elements (2873), MeJA-responsiveness elements (934), ABA responsiveness elements (574), Anaerobic induction elements (351), Gibberellin responsiveness elements (228), Drought-inducibility elements (185) was observed in the majority of the 1R-and 2R-ZoMYB genes. The 3R-ZoMYB gene (ZoMYB#197) is notably enriched with light-responsive elements (15 in total) and possesses a modest array of stress-responsive elements, including two MeJA-responsiveness elements, two ABA responsiveness elements, and two anaerobic induction elements. The MSA-like element, associated with cell cycle, was uniquely present in 10 ZoMYB genes (ZoMYB#220, ZoMYB#44, ZoMYB#154, ZoMYB#89, ZoMYB#18, ZoMYB#102, ZoMYB#68, ZoMYB#27, ZoMYB#26, ZoMYB#74). A quartet of meristem expression elements were identified in each of ZoMYB#186, ZoMYB#133, and ZoMYB#25. Furthermore, a singular MYB binding site, integral to the regulation of flavonoid biosynthetic gene elements, was found within ZoMYB#205, ZoMYB#124, ZoMYB#207, ZoMYB#147, ZoMYB#88, ZoMYB#117, ZoMYB#65, ZoMYB#132, ZoMYB#48, ZoMYB#24.
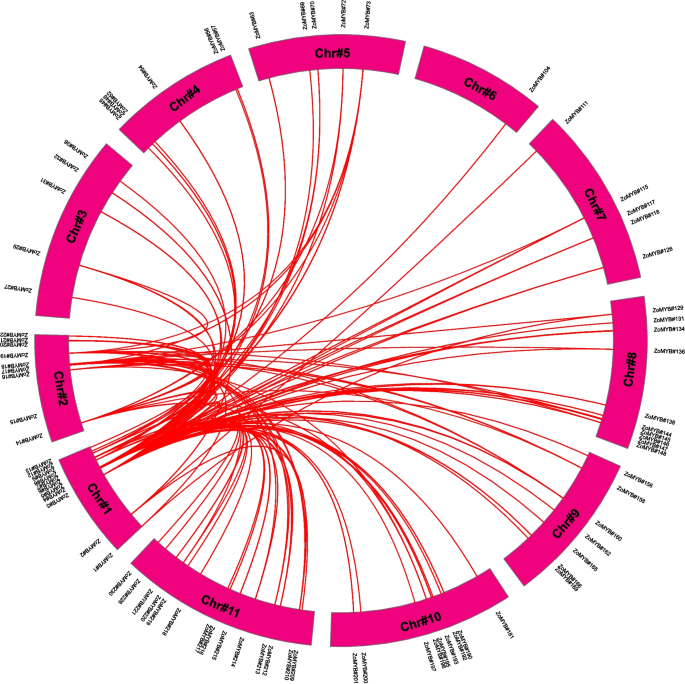
Chromosomal distribution, gene duplication, and synteny analyses of ZoMYB genes
Chromosomal allocation studies revealed a heterogeneous dispersal of ZoMYB genes across the 11 chromosomes of ginger (Supplementary Figure S4). Chromosomes 10, 6, and 8 harbored the highest contingents of ZoMYB (29, 27, and 27 genes, respectively), whereas chromosome 2 presented with a minimal assemblage of 9 ZoMYB genes. Proliferation of the ZoMYB gene family has predominantly been propelled by gene duplication events. It was found that 34 ZoMYB genes are clustered into 15 tandem duplication regions among ginger chromosomes 4, 5, 6, 8, 9, 10, and 11(Supplementary Figure S4 and Supplementary Table S4). Three tandem-duplicate gene pairs were found on chromosome 6, (ZoMYB#87-ZoMYB#88, ZoMYB#93-ZoMYB#94, ZoMYB#89-ZoMYB#90) and chromosome 10 (ZoMYB#188-ZoMYB#189, ZoMYB#184-ZoMYB#185, ZoMYB#193-ZoMYB#194); Additionally, two tandem-duplicate gene pairs were detected on chromosome 5 (ZoMYB#66-ZoMYB#67, ZoMYB#75-ZoMYB#76), chromosome 8 (ZoMYB#147-ZoMYB#148, ZoMYB#138-ZoMYB#139-ZoMYB#140), chromosome 9 (ZoMYB#165-ZoMYB#166, ZoMYB#170-ZoMYB#171) and chromosome 11 (ZoMYB#225-ZoMYB#26, ZoMYB#210-ZoMYB#211), with a sole tandem-duplicate gene pair on chromosome 4 (ZoMYB#42-ZoMYB#43). Among these tandem duplicated gene pairs, eight exhibited identical (ZoMYB#66-ZoMYB#67, ZoMYB#75-ZoMYB#76) or homologous (ZoMYB#89-ZoMYB#90, ZoMYB#42-ZoMYB#43, ZoMYB#138-ZoMYB#139-ZoMYB#140, ZoMYB#147-ZoMYB#148, ZoMYB#87-ZoMYB#88, ZoMYB#184-ZoMYB#185) motifs. And the remaining seven pairs contained different motifs (ZoMYB#93-ZoMYB#94, ZoMYB#165-ZoMYB#166, ZoMYB#170-ZoMYB#171, ZoMYB#188-ZoMYB#189, ZoMYB#193-ZoMYB#194, ZoMYB#225-ZoMYB#226, ZoMYB#210-ZoMYB#211). Beyond tandem duplications, 82 pairs of segmentally duplications were found within the ginger genome (Supplementary Table S4). The synteny analysis highlighted a conservation of MYB transcription factors across ginger chromosomes, with numerous homologous genes situated on disparate chromosomal tracts (Fig. 5).
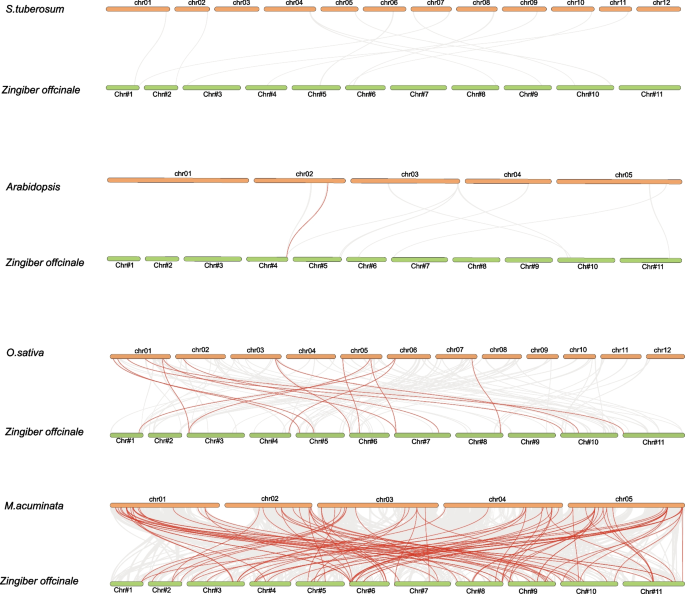
Evolutionary analysis of ZoMYB genes
In order to gain a deeper understanding of the phylogeny of the MYB family, we conducted a comprehensive synteny analysis comparing the ginger genome with those of four divergent plant species, including two monocots (M.acuminata and O.sativa), and two dicots (A. thaliana and S. tuberosum). The analysis revealed that a total of 75 ZoMYB genes exhibit syntenic relationships with 91 corresponding genes in M.acuminata, This was followed by 14 syntenic relationships with O.sativa, and a single syntenic relationship with an A. thaliana gene (Fig. 6 and Supplementary Table S5). There were 91, 15, and 1 pairs of orthologous genes identified between ginger and M. acuminata, O. sativa, and A. thaliana, respectively. ZoMYB#153 (Maker00037045) was found to be syntenic with three gene pairs in M.acuminata, while ZoMYB#216 (Maker00052236) corresponded with four M. acuminata gene pairs. The ZoMYB genes demonstrated a significant degree of orthology with the reference genomes, with M.acuminata demonstrated a significant degree of orthology with the reference genomes, following by O.sativa which displayed 15 orthologous gene pairs scattered across chromosomes 1, 2, 3, 5, and 7, and A. thaliana, with a single orthologous gene pair on chromosome 2. However, no syntenic relationships were identified between ginger and S. tuberosum (Fig. 6 and Supplementary Table S5). Further examination of the syntenic relationships between ginger and M. acuminata MYB genes revealed that 10 ZoMYBs were linked to two syntenic gene pairs each, 2 ZoMYBs were identified to be associated with 3 syntenic gene pairs each, and 1 ZoMYB gene was linked to 4 syntenic gene pairs.
To enrich our comprehension of the evolutionary pressures imposed upon the MYB gene lineage, an analysis of the nonsynonymous to synonymous substitution rate ratio (Ka/Ks) for MYB gene pairs was undertaken. As delineated in Supplementary Table S6, it was observed that the Ka/Ks ratios for all tandemly and segmentally duplicated ZoMYB gene pairs, as well as the majority of orthologous MYB gene pairs, were discerned to be less than one, indicative of purifying selection acting upon these sequences.
Expression profiling of ginger ZoMYB genes in different tissues
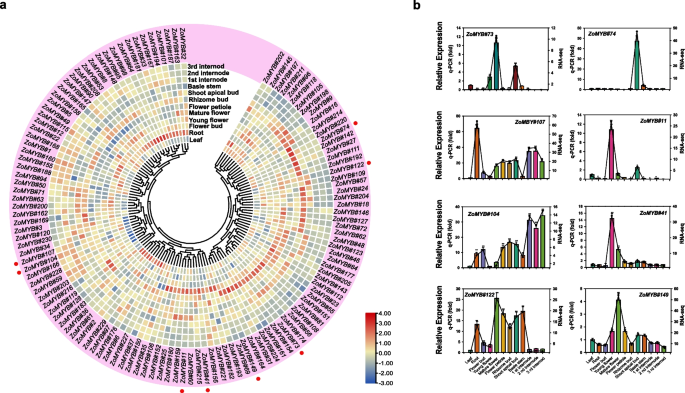
To explore the potential function of the ZoMYB genes at different stages of ginger development, RNA-sequencing data were utilized to ascertain their expression profiles (Fig. 7a and Supplementary Table S7). To further validate the reliability of the transcriptome data, qRT-PCR analyses were carried out on 12 representative samples for 12 selected ZoMYB genes (Fig. 7b). Among the 231 ZoMYB family genes, three ZoMYBs (ZoMYB#92, ZoMYB#94, ZoMYB#100) were not detected in any tested samples, suggesting the possibility of highly specialized spatio-temporal expression patterns that our dataset did not capture, or alternatively, these may represent pseudogenes. Among the 12 samples tested, the expression of 100 ZoMYB genes were detected (FPKM > 0), and 26 ZoMYB genes were constitutively expressed (FPKM > 1 in all samples). Eight ZoMYB genes (ZoMYB#98, ZoMYB#53, ZoMYB#208, ZoMYB#213, ZoMYB#145, ZoMYB#167, ZoMYB#101, and ZoMYB#95) are preferentially expressed in roots, one gene in flower bud (ZoMYB#12), four in young flower (ZoMYB#41, ZoMYB#11, ZoMYB#152, ZoMYB#215), two in mature flower (ZoMYB#149 and ZoMYB#73), three in shoot apical bud (ZoMYB#74, ZoMYB#72 and ZoMYB#66), one in leaves (ZoMYB#227) shown relative higher expression level than other ZoMYBs. The expression pattern of some ZoMYBs exhibited divergent tendency during different stage of development. For example, the expression levels of ZoMYB#143/182/155/202 were gradually increased, whereas that of ZoMYB#34/230/61/161/22/23/158/162 were gradually decreased during the rhizome development stages (Fig. 7a). Eight ZoMYBs were randomly selected to validate RNA-seq result by qRT-PCR. The results showed that the results are consistent with the RNA-seq (Fig. 7b).
ZoMYB gene expression profile analysis in ginger. a Hierarchical cluster analysis of ZoMYB gene expression profiles in 12 different tissues and developmental stages of ginger as determined by RNA-seq. The red circles represent the randomly selected ZoMYB genes that were randomly selected for validation of their expression profiles by qRT-PCR. b The expression levels of 8 ZoMYB genes were analyzed in 12 samples using qRT-PCR and RNA-seq. The X-axis represents the various tissues, while the Y-axis displays the q-PCR fold changes(vertical bar) and FPKM values(line and scatter) of the candidate genes on the left and right sides, respectively. Data are normalized to the Tub-2 gene, vertical bars are standard bar. n = 3. Mean values and standard deviations (SDs) were obtained from three biological and three technical replicates. The error bars indicate standard deviation. **P < 0.01 and *P < 0.05
Expression profiles of ZoMYB genes under abiotic stress conditions
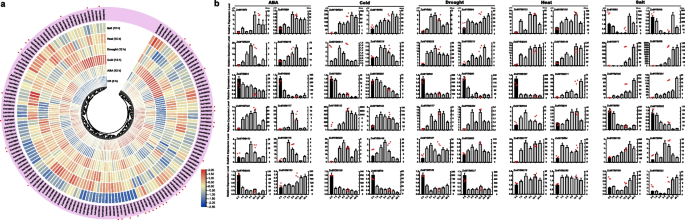
To elucidate the potential roles of ZoMYB genes under various abiotic stresses, we analyzed RNA-seq data to determine their transcriptional responses to heat, cold, salt, drought, and ABA treatments. A total of 154 ZoMYB genes were induced or reduced in response to at least one stress treatment compared with time 0 (CK) (Fig. 8a). Concisely, exposure to cold stress prompted the upregulation of 39 ZoMYBs, drought stress influenced 37, heat stress affected another 37, ABA treatment modulated 48, and saline conditions altered the expression of 62 genes (Supplementary Table S8). A subset of 8, 1, 4, and 9 ZoMYBs exhibited unique enhancements in response to ABA, cold, heat, and salt stress, respectively. Notably, drought stress did not uniquely induce any ZoMYB gene. A total of 13 ZoMYBs exhibited an upregulated expression pattern across all five abiotic stress treatments. (Supplementary Figure S5). Ten of these 13 ZoMYBs are R2R3-MYB types, while the remaining three are MYB-related types. These include three genes from the Z34 group, three from Z18 group, and two from Z25 group. ZoMYB#51 was enhanced in drought, heat and ABA stress treatment. Conversely, 49, 32, 40, 25, and 24 ZoMYB members were down-regulated by cold, drought, heat, ABA, and salt treatments, respectively. Ten ZoMYBs were reduced in all the five abiotic stress treatment, with four belonging to the Z34 group (R2R3-MYB type) and the Z10 group (MYB-related type). Distinct decreases were observed in 3, 10, 4, 1, and 1 ZoMYBs in response to ABA, cold, heat, drought, and salt stress, respectively. (Supplementary Figure S6). Nine ZoMYBs were reduced in both drought and heat stress. Among these differentially expressed genes, ZoMYB#3, ZoMYB#177, ZoMYB#221, belong to Z34 group were induced, while four genes, namely ZoMYB#60, ZoMYB#5, ZoMYB#105, and ZoMYB#202 also belong to Z34. Furthermore, we evaluated the expression patterns of 41 randomly selected ZoMYB genes under heat, low temperature, drought, salt and ABA stress conditions using qRT-PCR. Each of the 41 chosen ZoMYB genes manifested significant upregulation at one or several time points during stress exposure (as illustrated in Fig. 8b), corroborating the expression trends observed in the RNA-seq analysis.
Expression profiles of ZoMYB genes under various abiotic stress treatments. a ZoMYB genes expression in response to various abiotic stress as determined by RNA-seq. Relative expression of different ZoMYBs is shown for control and under ABA, cold, drought, heat, and salt stress after 12 h. Red circles indicate the ZoMYB genes that were arbitrarily chosen for validation of their stress-induced expression changes through qRT-PCR. b The expression of ZoMYB genes under abiotic stresses was analyzed using q-PCR and RNA-seq. The X-axis represents different time points after treatment, while the Y-axis displays qRT-PCR fold changes(left, vertical bar) and Fragments Per Kilobase Million (FPKM) values (right, scatter) of candidate genes. The expression levels were normalized to TUB-2 gene, and vertical bars indicate ± SD. n = 3. Mean values and standard deviations (SDs) were obtained from three biological and three technical replicates. The error bars indicate standard deviation. **P < 0.01 and *P < 0.05
Subcellular localization of ZoMYB proteins
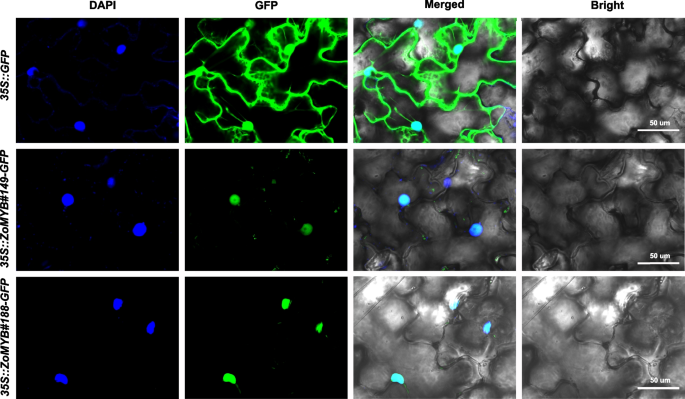
The subcellular localization of ginger MYB proteins from ginger was initially predicted using WoLF PSORT (Supplementary Table S1). Among the ZoMYB proteins, a substantial majority of 90.48% (209/231) were predicted to to localize to the nucleus. Additionally, 5.19% (12/231) were predicted to be chloroplastic, 1.30% (3/231) mitochondrial, another 1.30% (3/231) peroxisomal, 0.865% (2/231) cytoplasmic, and the remaining 0.865% (2/231) cytoskeletal. To verify these predictions, two genes, ZoMYB#149 and ZoMYB#188, which exhibited pronounced responsiveness to elevated temperatures and were specifically expressed in certain tissues, were selected for empirical validation through a transient expression assay. The fusion proteins ZoMYB#149-GFP and ZoMYB#188-GFP were observed to accumulated in the nuclei of epidermal cells. This was in stark contrast to the control GFP alone (vector control, 35::GFP), which was distributed throughout both the cytoplasm and the nucleus of the epidermal cells (Fig. 9). The empirical findings were congruent with the predicted subcellular localizations, reinforcing the accuracy of the initial predictions.
Subcellular localization of seven GFP-fused ZoMYB proteins. Transformation of tobacco epidermal cells with vectors containing 35S::ZoMYB-GFP constructs or a 35S::GFP control was followed by fluorescence examination via confocal microscopy at 40 h post-transfection.. Nuclear targeting was confirmed with DAPI staining. Scale bars = 50 μm







Add Comment