N. fowleri analysis under wet film observation was positive for the enflagellation test. Other morphological observations showed amoebic/cyst form, from CSF and culture. Both of these were considered for proper identification of the pathogen. Protruding pseudopodia was observed in fresh CSF wet preparation (Supplementary file 1.mp4). Further, to confirm Naegleria fowleri, 18 S-ITS1-5.8 S-ITS2-28 S region of 410 bp was amplified using PCR as shown in figure-1.
N. fowleri CERE assembly and annotation
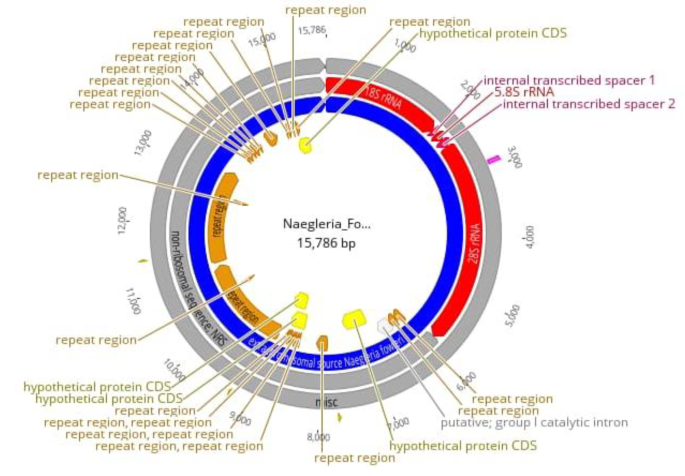
CERE-rDNA sequence data was quality-filtered for further downstream processing. High-quality assembly reads were first aligned against N. fowleri (CM017919.1) followed by the assembly of the CERE – rDNA genome. The major features of CERE elements include rRNA genes, repeats, and ORFs. The CERE-rDNA size of N. fowleri Karachi isolate that we sequenced is up to 15.79 kb having 40.5% GC content (Figs. 1 and 2). However, the previously identified CERE-rDNA from different Nagleria spp shows some variation in its size; The CERE-rDNA size is 15.79 kb in N. fowleri strain LEE MT741533.1, 15 kb in N. gruberi, 14 kb in N. lovaniensis, 15 kb in N. jamiesoni, 13.6 kb in N. australiensis and 11.8 kb in N. jadini [3,4,5,6]. Most of the size difference in CERE elements of different Naegleria specie is due to the variation in their non-rDNA sequence (NRS); however, their rDNA sequence have almost similar size having only minor differences in the internal transcribed spacers (ITS) [16].
The 18 S in CERE element comprised of 2027 bp followed by two internal repeats of 223 bp separated by 144 bp 5.8 S. The 28 S rRNA comprised 3465 bp followed b y repeat elements and hypothetical proteins. Repeat elements comprised 7268 bp (46.04) in the whole CERE element.
Our N. fowleri isolate was compared with other N. fowleri isolates to assess their evolutionary relationship and single nucleotide polymorphisms (SNPs), along with insertions and deletions. Variants with a quality score of less than 30 were removed. Variants showed higher variability among various N. fowleri isolates analyzed in this study. There were 90 variants in total, including 41 variants in 18s rRNA and 49 variants in 28s rRNA region of CERE – rDNA. N. fowleri strain LEE MT741533.1 showed a deletion of 44 nucleotides at the 2026 position of 18s rRNA, and T to A transition on direct repeat region at position 8040. N. fowleri CM017919.1 had an insertion in tandem repeat at position 6,207 of about 167 nucleotides. The second insertion was of 22 nucleotides as a direct repeat at position 14,982. A deletion (direct repeat from ACCC to ACC at position 12,354) was also seen. Besides these insertions and deletions, 11 SNPs were also present (Supplementary file 2).
Phylogenetics and recombination events
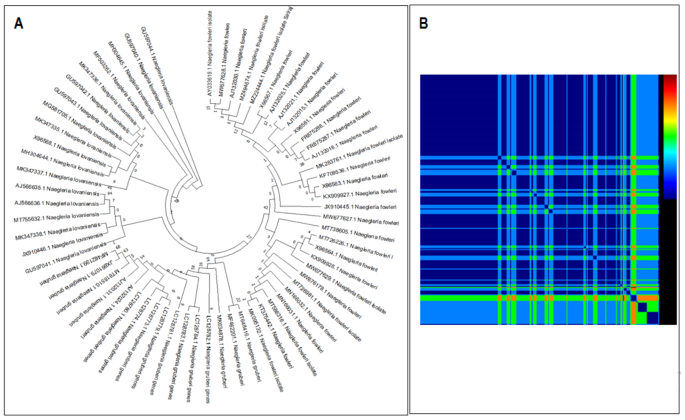
Our isolate and N. fowleri (CM017919.1), N. fowleri strain LEE (MT741533.1), N. fowleri Karachi_NF001 strain (OD958550.1), N.gruberi (AB298288.1), and N. lovaniensis (CM010402.1) were studied using Neighbor-Joining method to evaluate evolutionary relationship across various CERE – rDNA. The ITS-I DNA sequences from all CERE- rDNA of different species included here for phylogenetic analyses showed different patterns of evolutionary relationship (Fig. 3). CERE-rDNA DNA sequences of N. fowleri Karachi isolate showed maximum similarity with N. fowleri strain LEE and N. lovaniensis CERE-rDNA sequence also showed close homology. N. gruberi formed a separate group, while N. fowleri Karachi_NF001 and N. fowleri species(CM017919.1) were forming a separate clade. This observation is quite interesting because all CERE-rDNA sequences used for phylogeny analyses belong to separate species. Hence a regularly used internal transcribed spacer I and 5.8 S ribosomal RNA genes were considered for further phylogenetic analysis. The phylogenetic tree was constructed using 66 sequences. It showed that the pattern of evolution and clade formation was different for different species (Fig. 4A). These analyses indicate that ITS-I, ITS-II, and 5.8 S rRNA are of great diagnostic value for rapid amoeboid identification and differentiation. The Karachi isolate CERE – rDNA showed a different pattern in the NJ tree, this could be due to low sample size as a low number of CERE – rDNA have been reported so far. A total of 22 recombination events were predicted and these were screened for actual recombination events (Fig. 4B). The analyses resulted in some over-expressed sequences and were subsequently eliminated. Further stringency was increased by considering parent recombination events (from both major and minor parental recombinations), identified by their presence in both sequences. Among 5 recombination events, three were found among N.gruberi (AB298288.1) and N. fowleri strain LEE (MT741533.1) CERE – rDNA, starting from 9,366 to 9,754 bp,13,485 to 13,772 bp, and 11,892 to 11,994 bp, respectively. N.gruberi (AB298288.1) and OD958550.1 showed two recombination sites (23,461–24,449) while other recombination events occurred between N.gruberi (AB298288.1) and N. fowleri strain LEE (MT741533.1) (22,649–23,287) (Supplementary file 3). These recombination events could explain possible variability among the different patterns in terms of genetic heterogeneity (Fig. 3).
Hypothetical protein structures and functionality evaluation
Four hypothetical proteins were studied for their physiological and biochemical analysis. The hypothetical protein 4 (Hypo-4) containing 104 amino acids, showed a molecular weight of.
10552.20 Dalton, theoretical pI: 11.63, and Grand Average of Hydropathicity (GRAVY): 0.284. Hypo-4 protein was classified as stable with an estimated half-life of 20 h (> 20 h in yeast having the instability index (II) computed to be 11.15) [38].
Protein structure and model quality assessment
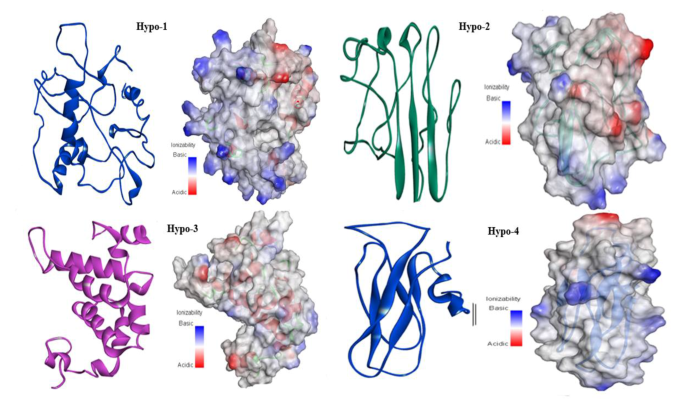
All hypothetical proteins including Hypo-1, Hypo-2, Hypo-3, and Hypo-4 were studied for their proper secondary structures using PSIPRED, SOPMA, and ENDscript servers. In three hypothetical proteins, the random coil was the most predominant feature with 53.85%, 49.30%, and 60.58% occurrence in Hypo-1, Hypo-2, and Hypo-4, respectively, while in Hypo-3, it was only 20.55%. Hypo-2 and hypo-4 proteins belong to all b-class of protein folds and are found to consist of 12.68% and 13.46% b-elements, respectively. ENDscript and PSIPRED showed similar results. The Models were predicted by I-TASSER and checked for proper structure prediction using I-TASSER scoring. The structure for Hypo-1 proteins lacks regular secondary structure, although its fold has few a-helices. The majority of the surface area showed basic potential, localized on one side of the protein which may be involved in nucleotide binding. This protein was predicted to have ATP binding/ligase activity. The Hypo-2 protein (Fig. 5) had 50% loop region while the other 50% consisted of b-sheets having three b-strands, representing the mixed type of surface potential. It was predicted to have ribonuclease-inhibitor activity. The Hypo-3 protein is included in all a-class of proteins. It has ATP binding/glucosidase activity. The Hypo-4 protein belongs to all b-class of proteins and is predicted to contain hydrolase activity against the O-glycosyl compounds e.g., polysaccharides. The surface representation of this protein depicts the overall basic surface potential which facilitates the binding of this protein with polysaccharide.
Protein binding site and virtual screening
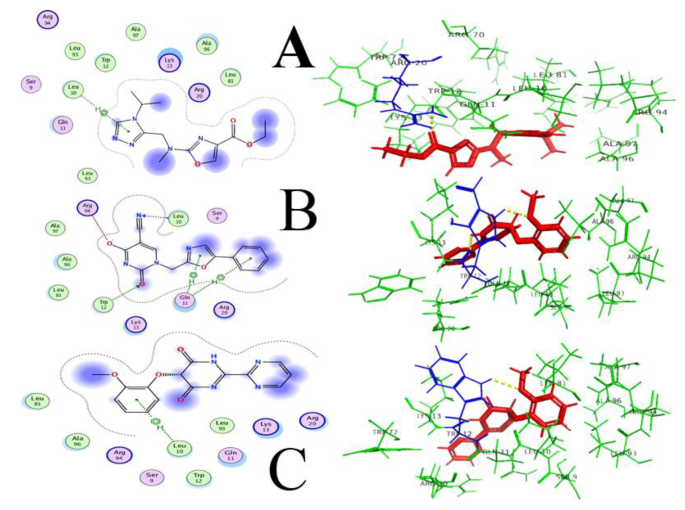
As Hypo-4 depicted a proper conformation and structure, it was selected for further analysis. Domain conservation analysis along with functional annotation and functional site identification was performed using BLASTp search, NCBI-CD Search, Pfam, and InterProScan. The motor domain of Dictyosteliumdiscoideum cytoplasmic dynein (PDB ID: 3VKG) was selected as a template that showed similarity to hypo-4 protein, having 15% query coverage and 68.75% sequence identity with an E-value of 0.020. The structural superimposition was performed to check the structural similarity and active site comparison in both structures, where adenosine-5’-diphosphate ligand was bound nearby of the template structure. This ligand structure was used for pharmacophore-based screening (11 features selected in MOE software), with a ZINC library of 11,193 drug-like molecules. 1,271 compounds gave the best hits in virtual screening and the top 20 hits (Supplementary file 4) were tabulated considering their S-value and were further subjected to ADME analyses (Administration, Distribution, Metabolism, and Excretion) and toxicity. Compounds with ID: ZINC77564275, ZINC48229542, and ZINC15022129 were selected as potential drugs based on their S-score and interaction parameters (Fig. 6). These top compounds having higher binding affinities were then analyzed for pharmacokinetics and pharmacodynamics using their administrant, distributions, metabolism, and excretion (ADME) profile. Compounds showing blood-brain permeability should be key for considering pathogen that resides in the brain for its pathogenesis, as drug molecules need to cross blood blood-brain barrier. All compounds were predicted substrates of P-glycoprotein, showing no inhibitory effect against the cytochromes CYP3A4, CYP2C9, CYP2C19, CYP2D6, and CYP1A2. No compound depicted potential carcinogenicity while ZINC48229542 was positive for Ames mutagenesis, and probably not be a suitable drug.
ZINC77564275 (ethyl 2-(((4-isopropyl-4 H-1,2,4-triazol-3-yl)methyl)(methyl)amino)oxazole-4-carboxylate) and ZINC15022129 (5-(2-methoxyphenoxy)-[2,2’-bipyrimidine]-4,6(1 H,5 H)-dione) were finalized, based on ADME toxicity analysis. These two compounds showed least to no toxicity to the host and honey bees, crustacea, and fish. Selected final compounds were also positive for biological degradation, and safe for the environment. To the best of our knowledge, the proposed compounds are novel and safe, as they have not been previously reported in the literature for their anti-Naegleria-like activities.





Add Comment