System overview
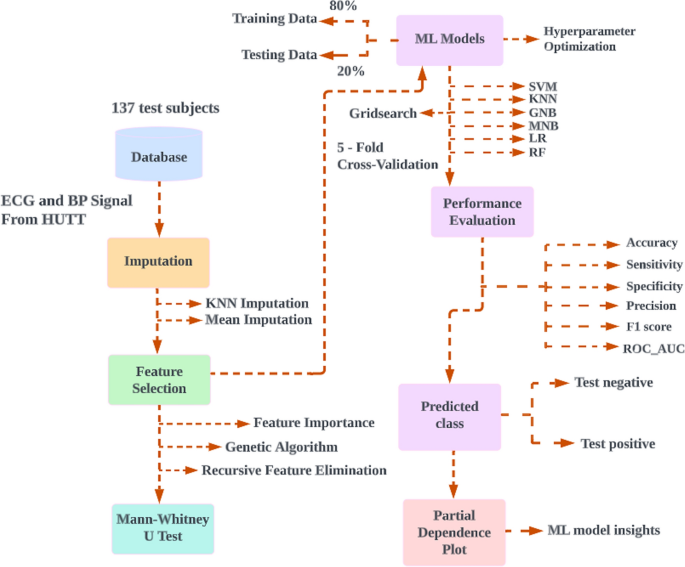
The proposed method was developed with a laptop equipped with Intel (R) Core (TM) i7-6600U 2.60 GHz CPUs and 8 GB of RAM. The python packages that were used included numpy, pandas, scikit-learn, matplotlib, and seaborn. Data collection, experimental setup, data preprocessing on physiological signals, selection of features, missing value imputation, statistical analysis, ML classifiers, and PDP is all demonstrated in this section. Figure 4 represents the proposed model for classification of VVS.
The proposed model for classification of vasovagal syncope. A flow diagram showing the proposed model for classification of vasovagal syncope. Features ECG and blood pressure signals from 137 HUTT were first extracted then imputed. Selected features identified using feature selection methods and non-parametric probability testing were performed in order to compare the statistical differences between two groups then cross validated and 80% of the data used as training set for machine learning, with the remaining 20% as testing set. The model performance was evaluated, then prediction classification and partial dependence plot applied. HUTT, head-up tilt test; ML, machine learning; SVM, support vector machine; K-nearest neighbors (KNN) imputation dataset; GNB, Gaussian naïve Bayes; MNB, multinomial naïve Bayes; LR, logistic regression; RF, random forest
Data collection
Data collection was conducted at the Cardiorespiratory Laboratories, University of Malaya Medical Centre (UMMC). Patients were referred for HUTT as an investigation for the symptom of syncope or near syncope. All patients were provided information about the test and informed consent obtained prior to the test. Both the Universiti Tunku Abdul Rahman (UTAR) Scientific and Ethical Review Committee (U/SERC/218/2020) and the UMMC Medical Research Ethics Committee (MREC ID NO: 2,020,913–9066) provided their approval for this cross-sectional study.
Experimental setup
The head-up tilt test was meticulously conducted, employing specialized non-invasive monitoring equipment furnished by CNSystem, with a central focus on the Task Force™ Monitor (CNSystems, Graz, Austria). This sophisticated monitoring system was purpose-built to facilitate comprehensive hemodynamic measurements, enabling seamless real-time data acquisition and subsequent analysis [27]. Core bio signals, comprising ECG and BP were meticulously recorded on a beat-to-beat basis. It is worth mentioning that the Task Force Monitor was carefully outfitted with a high-resolution two-channel ECG module, proficiently capturing data at a frequency of 1000 Hz. Additionally, a BP module, operating at 100 Hz, further augmented the system’s capabilities. Subjects underwent meticulous preparation, entailing the precise attachment of electrodes and sensors to capture pivotal physiological parameters such as BP and HR. These well-prepared subjects were subsequently positioned on an integrated tilt table, thoughtfully designed to enable controlled angle adjustments. The tilt table served as a dynamic platform, allowing subjects to undergo a gradual transition from a supine position to a predetermined 70-degree angle. The protocol HUTT was carried out in a tranquil and carefully managed setting designed to minimize external factors. The substantive phase of the test commenced after a designated 10-min period of supine rest. The substantive phase of the test commenced after a designated 10-min period of supine rest. Subsequently, the tilt table was artfully maneuvered to achieve a consistent 70-degree angle, sustained total a period of 35 min [28]. Notably, a pharmacological stimulus was introduced after the initial 20 min of tilting, which involved administering 400 µg of glyceryl trinitrate (GTN) sublingually. This deliberate measure aimed to evoke specific physiological responses during the tilt phase. The outcomes of the test are determined based on a rigorous analysis of the observed physical alterations and their correlation with subjects’ prior symptoms during spontaneous incidents. Positive results ensue when discernible physical changes, such as diminished HR, cardiac interruptions, and reduced BP, precisely mirror patients’ past symptomatic episodes. In contrast, if physical changes manifest without replicating symptoms, the results are categorized as false positives. Conversely, negative results are established when symptoms fail to align with observed physical changes or when neither symptoms nor physical alterations manifest during the test [13].
Data preprocessing on physiological signals
Our research integrates various aspects to provide a comprehensive understanding of physiological responses during the study. To begin, the feature extraction approach entails the analysis of two distinct time periods. The initial 10-min segment positions subjects in a supine resting state, followed by a subsequent 15-min period during which they are tilted at a 70-degree angle with administered GTN. This detailed approach allows for a thorough analysis of beat-to-beat HR and BP signals.
In the context of the HUTT, the timing of VVS occurrence varies among subjects and situations. Typically, VVS manifests during the test itself, although the precise timing can differ. Some subjects experience VVS early in the HUTT window, within the first few minutes, while others encounter it later. These variations can be attributed to the unique physiological responses, medical history, and specific test circumstances of the subjects.
As part of the study, an extensive data preprocessing methodology for physiological signals was employed. This involved a thorough examination of time domain parameters of heart rate variability (HRV) and blood pressure variability (BPV). Additionally, an in-depth analysis of the frequency domain variability in heart rate and blood pressure was conducted. This multifaceted data preprocessing approach is crucial for obtaining a comprehensive understanding of the physiological responses throughout the study.
Time domain parameters of HRV and BPV
The modulation of HR involves both the sympathetic and parasympathetic branches of the ANS. Sympathetic activity increases HR but lowers HRV, while parasympathetic activity decreases HR but increases HRV [29]. The regulation of autonomic output involves interconnected parts of the Central Nervous System (CNS). In certain situations, the vagus nerve can react abnormally, causing a sudden drop in BP, slower HR, widened blood vessels, excessive sweating, and various symptoms like dizziness, nausea, blurred vision, ultimately leading to fainting (syncope) [6,7,8]. Moreover, fluctuations in BP result from complex interactions between various cardiovascular systems, including central autonomic regulation, sympathetic vascular modulation, baroreflex, and humoral influences [30]. In response to PNS activation, BP is lowered through vasodilation and bradycardia to prioritize blood flow to essential organs [8]. However, in VVS, these normal responses are disrupted or magnified, leading to an excessive drop in BP and HR due to factors such as an overactive PNS, heightened blood vessel sensitivity, and abnormal autonomic signalling [10]. When it comes to properly assessing the contributions of multiple underlying regulatory mechanisms, time-domain metrics reflecting the entire variability of both HR and BP are rather indiscriminate. The study calculated the standard deviation (SD), average real variability (ARV), root mean square of real variability (RMSRV), coefficient of variation (CV), and standard deviation of real variability (SDRV) in a study of both HRV and BPV in the time domain. These indexes’ formulas are listed below [31]:
$$\mathrm{Standard\; deviation }\left({\text{SD}}\right)=\sqrt{\frac{{\sum }_{i=1}^{n}{\left({x}_{i}- \overline{x }\right)}^{2}}{n-1},}$$
(1)
$$\mathrm{Coefficient\; of\; variance }\left({\text{CV}}\right)=\frac{SD}{mean} x 100\%.$$
(2)
Average real variability (ARV): Variations in absolute terms between the subsequent values:
$$=\frac{\sum_{i=1}^{n-1}{D}_{i}}{n-1}, where\; {D}_{i=\left|{x}_{i+1}- {x}_{i}\right|.}$$
(3)
Root mean square of real variability (RMSRV): The sequential difference between the subsequent values expressed as the root mean square:
$$=\sqrt{\frac{{\sum }_{i=1}^{n-1}{({D}_{i})}^{2}}{n-1}.}$$
(4)
Standard deviation of real variability (SDRV): Sequential differences between the surrounding values:
$$=\sqrt{\frac{{\sum }_{i=1}^{n-1}{\left({D}_{i}- \overline{D }\right)}^{2}}{n-1}.}$$
(5)
Here, x = HR, SBP, and DBP are all measured from beat to beat, \(\overline{x }=\) mean for the relevant variable and n = the overall number of beats for the selected variable.
Frequency domain variability in heart rate and blood pressure
The spectrum analysis plots the variation of spectral power of HRV and BPV as functions of frequency. The adaptable autoregressive coefficients generated with each physiological parameter were utilized to calculate the frequency spectrum in the Task Force Monitor [32]. The following parameters can be determined via spectral analysis:
LF scale 0.04–0.15 Hz [33],
HF scale 0.15–0.4 Hz [34].
$$\mathrm{LF\; normalized\; units}-\mathrm{LF\; n}.{\text{u}}. =\frac{LF}{LF+HF}*100,$$
(6)
$$\mathrm{HF\; normalized\; units}-\mathrm{HF\; n}.{\text{u}}.=\frac{HF}{LF+HF}*100.$$
(7)
Since HF + LF < 100.
The relationship and balance of both sections of the ANS can be determined via spectral analysis, however, absolute levels of HRV and BPV features are not indicators of ANS activity. The HF domain is thought to be mostly influenced by parasympathetic modulation, while the LF is primarily influenced by sympathetic modulation. The LF-to-HF ratio is used to estimate the balance of both components of the ANS’s influence on the heart.
Missing value imputation
In our study, which encompassed data from 137 subjects undergoing HUTT, both test-positive and test-negative subjects were included. Within this group, we identified missing data in a total of 17 subjects—comprising 12 test-negative and 5 test-positive subjects. The specific missing data pertained to features referred to as systolic blood pressure variability in both supine and tilting positions [Hfnu_SBPV (high-frequency normalized power of systolic BP variability), Lfnu_SBPV (low-frequency normalized power of systolic BP variability), LFHF_SBPV (ratio of high frequency to low frequency normalized power of systolic BP variability)] and diastolic blood pressure variability in the same positions (Hfnu_DBPV (high-frequency normalized power of diastolic BP variability), Lfnu_DBPV (low-frequency normalized power of diastolic BP variability), LFHF_DBP (ratio of high frequency to low frequency normalized power of diastolic BP variability) within our dataset. Notably, the pattern of missing data we encountered was not at random. Interestingly, the same set of features exhibited missing data across all 17 subjects, suggesting a non-random and potentially systematic underlying cause for these gaps. These instances of missing data, often represented as “NAN” (Not a Number) values, can be attributed to several contributing factors, which may include cuff-related issues, variations in sensor sensitivity, technical anomalies, and signal saturation. These circumstances can lead to intermittent temporal gaps within our recorded data. It is important to highlight that these factors collectively influence the overall quality and comprehensiveness of our dataset, and they have the potential to impact subsequent analyses and interpretations. Confronted with the presence of “NAN” data during the HUTT, the medical experts chose to move forward, leveraging their clinical expertise. Following a thorough assessment of the situation, they determined that the data gap’s impact on the test’s reliability was minimal, taking into account considerations such as the patient’s health, the test’s objectives, and the potential consequences of restarting the test. This decision was bolstered by a robust 10-min supine baseline, a carefully controlled 70-degree tilt maintained for 20 min, and the introduction of a 400 µg GTN potentiation. In essence, their choice exemplified a commitment to patient-centered care, opting to avoid an early test restart in order to prioritize the well-being of the patient.
In this study, missing data were addressed using two methods: KNN imputation and mean imputation. For KNN imputation, implemented with Scikit-learn’s KNNImputer function, missing values were determined based on the Euclidean distance to the nearest neighbors. Mean imputation involved using the mean value of each feature, calculated through Python’s panda’s library, to fill in missing values. Both techniques were selected for their simplicity, efficiency, and widespread use. While KNN imputation captures underlying relationships for accuracy, mean imputation is widely adopted due to its ease of implementation [35]. By employing both methods, the study enabled performance comparison and a robust approach to managing missing data [36, 37].
Selection of features
Three distinct methods were employed to identify the most significant features within the dataset: FI, GA, and RFE. Independently, all three techniques were utilized, retaining features chosen by the most fitting candidates. For assessing feature importance (FI), the Scikit-learn library’s RF approach was adopted. Utilizing the featureimportances attribute, relevance scores were computed for each feature. The top three features were then identified for further analysis, following an averaging process.
The genetic algorithm (GA) approach utilized the LR classifier from Python’s Scikit-learn module, with parameters like crossover and mutation probabilities optimized for optimal feature selection. The fitness function was tailored to prevent overfitting, selecting relevant features based on cross-validation accuracy.
In recursive feature elimination (RFE), the decision tree algorithm from Scikit-learn was employed. With configuration to retain three features, the iterative process recursively eliminated the least significant feature, leaving behind the remaining features for in-depth examination.
Statistical analysis
To ascertain if test positive and test negative are subsets from the same population, the MWUT, a statistical hypothesis test, is performed. An affirmative outcome for a HUTT occurs when symptoms of syncope reoccur alongside a corresponding reduction in HR or BP. Conversely, a negative result pertains to the absence of symptom recurrence, regardless of the HR or BP changes, or when symptoms manifest without a corresponding HR or BP response. Test positivity was determined by the medical expert supervising the HUTT. If the statistical result exceeds 0.05 (P ≤ 0.05), the study rejects the null hypothesis and draws the conclusion that the two samples did not come from the same population [15].
Machine learning classifiers
In the study, six machine learning classifiers, namely SVM, KNN, MNB, GNB, LR and RF for VVS categorization.
Support vector machine
The study in Support vector machine (SVM) algorithm accustomed the linear or radial basis function (RBF) kernel. C is a penalty parameter with values 0.1,1,10 and 100. Gamma is in the range of 0.1, 0.001, 0.001, and 0.0001.
K-nearest neighbors
The K-nearest neighbors (KNN) algorithm’s power parameter for the Minkowski distance metric is denoted by “p”. p and n_neighbors in this experiment had values of 1 to 10.
Multinomial naïve Bayes
When it comes to HUTT data, multinomial naïve Bayes (MNB) can be utilized to assess the probability of a patient experiencing VVS. Our study had fine-tuned parameter alpha values such as 0.001,0.01,0.1,0.5,1.0,10.0, and 100.0.
Gaussian naïve Bayes
When working with continuous data, it’s common to assume that each class’s continuous values will be distributed in a Gaussian naïve Bayes (GNB). Here the range of var_smoothing was log space (0, − 9, number = 100).
Logistic regression
The frequency of a target attribute is forecasted using a LR. Here log space (-3,3,7) and 11 and 12 were the hyperparameters C and penalty, respectively.
Random forest
Due to the random selection of features, which lowers the correlation between the ensemble’s trees, this strategy tends to increase the ensemble’s predictive ability. The random forest (RF) model’s hyperparameters included max_depth ranges of 2 to 10, min_samples_leaf ranges of 5 to 200, and n_estimators range of 10 to 200.
Performance evaluation
The parameters of the models our study generated were altered using GridSearchCV with a fivefold cross-validation for fine-tuning all classifiers. The study employed ML algorithms, all of which were initialized with a random state value of zero. This approach aimed to ensure the reproducibility of results by generating the same sequence of random numbers in each run of the algorithms. The following formulas were used to calculate our proposed models:
$${\text{Accuracy}}=\frac{{\text{TP}}+{\text{TN}}}{{\text{TP}}+{\text{TN}}+{\text{FP}}+{\text{FN}}},$$
(8)
$$\mathrm{Recall }\left({\text{\;sensitivity}}\right)=\frac{{\text{TP}}}{{\text{TP}}+{\text{FN}}},$$
(9)
$${\text{Specificity}}=\frac{{\text{TN}}}{{\text{FP}}+{\text{TN}}},$$
(10)
$${\text{Precision}}=\frac{{\text{TP}}}{{\text{TP}}+{\text{FP}}},$$
(11)
$${\text{F}}1\mathrm{ \;score}=2*\frac{{\text{Precision}}*{\text{Recall}}}{{\text{Precision}}+{\text{Recall}}},$$
(12)
where true positives (TP), false positives (FP), false negatives (FN), and true negatives (TN) were all included in the calculation of the confusion matrix. Regarding subjects undergoing testing, when a subject tests positive and the model accurately categorizes them as positive, they are labeled as TP. Conversely, if a subject tests negative but is mistakenly classified as positive by the model, they are referred to as FP. In the same vein, a subject with a genuine negative test result is denoted as TN. On the contrary, if a subject who tests positive is incorrectly categorized as negative, it falls under the FN classification.
Partial dependence plot
Partial Dependence Plot (PDP) showed the behaviors of the model and assisted in determining which characteristics had the most influence on the outcome of the decision-making process [38, 39]. To implement this XAI model, the study employed the Scikit-learn module in Python with a RF classifier, effectively fitting the model.





Add Comment